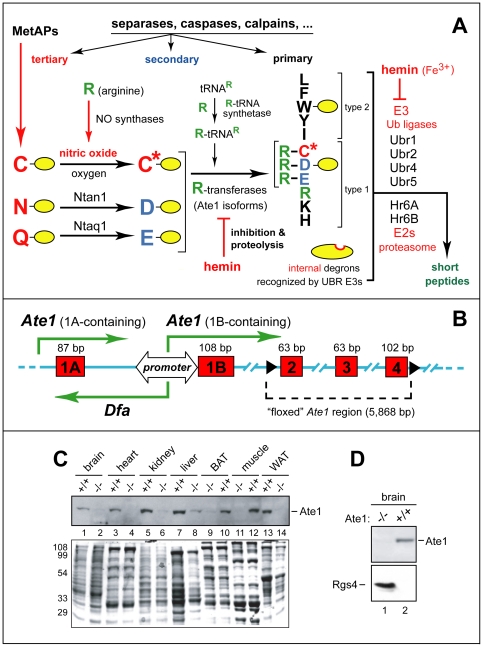
Figure 1. Postnatal ablation of the mouse Ate1 R-transferase, a component of the N-end rule pathway.
(A) The mammalian N-end rule pathway. N-terminal residues are indicated by single-letter abbreviations for amino acids. Yellow ovals denote the rest of a protein substrate. “Primary”, “secondary” and “tertiary” denote mechanistically distinct subsets of destabilizing N-terminal residues (see Introduction). C* denotes oxidized Cys, either Cys-sulfinate or Cys-sulfonate. MetAPs, Met-aminopeptidases. (B) Bidirectional promoter between the mouse Ate1 exons 1A and 1B [14]. Green arrows indicate transcriptional units, including a previously uncharacterized gene, termed Dfa (“divergent of Ate1), that is transcribed from the bidirectional promoter. (C) Immunoblotting-based comparisons of Ate1 levels in the indicated mouse tissues from Ate1+/+ and Ate1flox/−;CaggCreER mice 76 days after the tamoxifen (TM)-induced, Cre-mediated Ate1flox→Ate1− conversion that yielded Ate1-deficient mice. The band of 60-kDa Ate1, detected by antibody to mouse Ate1, is indicated on the right. Total (Ponceau-stained) protein patterns are shown below, with positions of molecular-mass markers on the left. (D) IB assays for the levels of Ate1 and Rgs4 (25 kDa) in brain extracts from Ate1+/+ and Ate1-deficient mice (Ate1flox/−;CaggCreER mice 30 days after TM treatment).