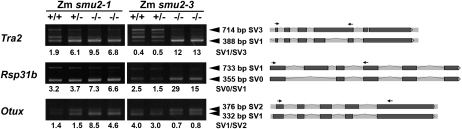
Figure 1.
Altered RNA splicing in Zmsmu2-1 and Zmsmu2-3 endosperm. Heterozygous (smu2-1/+ or smu2-3/+) plants were self-pollinated, and kernels were dissected into embryo and endosperm tissues at 16 d after pollination. Embryo DNA was used to genotype the Zmsmu2 locus, and RNA was extracted from homozygous wild-type, heterozygous, and homozygous mutant endosperms and used for semiquantitative RT-PCR. +/+, Endosperm with a homozygous wild-type embryo; +/−, endosperm with a heterozygous embryo; −/−, endosperm with a homozygous smu2-1 mutant embryo. Arrowheads indicate SVs with different band intensities between the wild type and mutants. Pre-mRNA splicing patterns of SVs are shown at right, with RT-PCR primers indicated by arrows. Ratios of differentially accumulated SVs are shown below the panels.