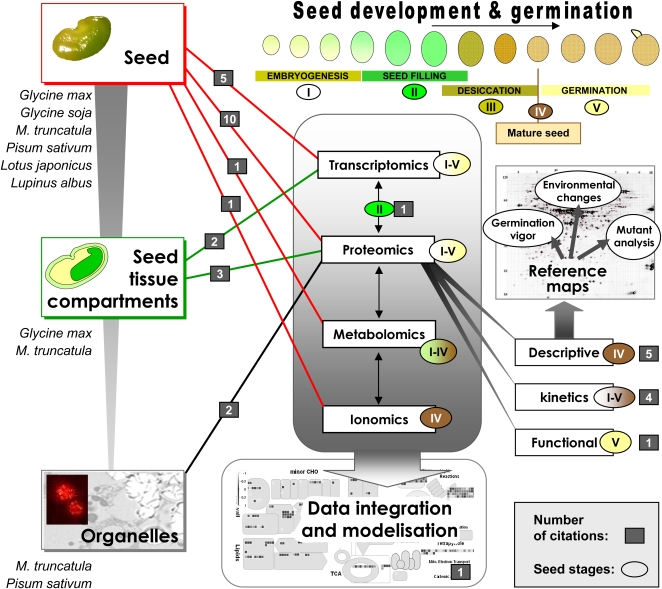
Figure 1.
Post-genomics strategies applied to legume seeds for dissecting their composition and/or identifying regulatory and metabolic networks during developmental processes. Gray squares represent the number of citations for each strategy. For the entire seed, the citations refer to post-genomics experiments carried out in various legume species at the level of the transcriptome (Firnhaber et al., 2005; Buitink et al., 2006; Gallardo et al., 2007; Benedito et al., 2008; Verdier et al., 2008), metabolome (Vigeolas et al., 2008), ionome (Sankaran et al., 2009), and proteome. At the level of the proteome, some citations refer to the mature seed (Watson et al., 2003; Magni et al., 2007; Bourgeois et al., 2009; Krishnan et al., 2009; Natarajan et al., 2009), to the kinetics of seed development (Hajduch et al., 2005; Gallardo et al., 2007; Agrawal et al., 2008; Dam et al., 2009), and to a functional study for identifying targets of thioredoxin (Alkhalfioui et al., 2007). For the separated seed tissues, the citations refer to transcriptome and proteome analyses in soybean (Le et al., 2007) and M. truncatula (Boudet et al., 2006; Zhang et al., 2006; Gallardo et al., 2007). For the seed organelles, the citations refer to proteome analyses of pea mitochondria (Bardel et al., 2002) and M. truncatula nuclei (Repetto et al., 2008). The citation referring to the comparison of transcriptome and proteome datasets is Gallardo et al. (2007) and that referring to data integration is Goffard and Weiller (2006).