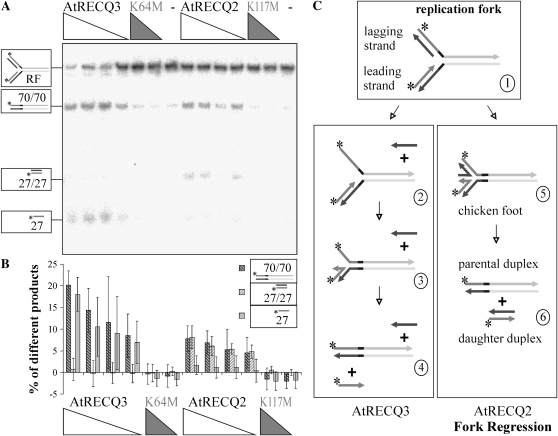
Figure 5.
Conversion of a synthetic replication fork (RF). A, Decreasing concentrations of AtRECQ2 and AtRECQ3 (8, 5, 4, and 2.5 nm) and AtRECQ2-K117M and AtRECQ3-K64M (8 and 5 nm) were incubated with 100 pm of the replication fork for 20 min. Helicase reaction products were analyzed on 12% native TBE polyacrylamide gels. On the left, the substrate and products are symbolized. Asterisks mark the 32P-labels. B, Quantification of the main products. The mean and sd of the results of seven experiments are shown. C, Proposed mechanisms of replication fork conversion by AtRECQ2 and AtRECQ3. In the scheme of the replication fork, identical sequences share a common color; complementarities can be deduced from the figure. On the left, the proposed mechanism for AtRECQ3 is shown. Unwinding of the lagging daughter strand enhances the chance of pairing the parental strands in the context of DNA breathing. This will lead to the displacement of the leading daughter strand. On the right, fork regression with the chicken foot intermediate is shown. Further branch migration leads to parental and daughter duplexes.