The major pest of soybean (Glycine max) is the soybean cyst nematode (SCN), Heterodera glycines. One population of SCN can evoke a resistant response while a second population can evoke a susceptible response from the same soybean cultivar. Recently, interactions between SCN and soybean roots have been studied using commercially available microarrays to measure both soybean and nematode transcripts. Furthermore, precise dissection of nematode feeding sites (syncytia) using laser capture microdissection (LCM) now allows the study of gene expression specifically in syncytia during a resistant and susceptible reaction. Genes and pathways that are up- and down-regulated in roots and syncytia during the interaction of soybean with SCN have been identified. In this Update, we discuss recent research on gene expression during interactions of soybean with SCN and how this information is being used to identify soybean and SCN genes involved in resistance and susceptibility.
BACKGROUND
Plant parasitic nematodes are a significant agricultural problem. Annually, they cause approximately 157 billion U.S. dollars in damage, worldwide (Abad et al., 2008). Plant parasitic nematodes can infect any part of a plant, both above and below ground. However, the most damaging and agriculturally relevant are the root parasites. Recent interest in soybeans, both as food and as a source of bioenergy, makes research into their pathogens of supreme interest to the agricultural industry. Plant parasitic nematodes present extensive challenges to the cultivation of soybeans. For example, there have been reports of over 20 genera of plant parasitic nematodes to be present in soybean fields (Sinclair and Backman, 1989). The major parasitic nematode for soybean is the SCN (H. glycines). This review focuses on recent scientific advances in the area of genomics and gene expression in relation to soybean interactions with SCN.
SCN is an obligate sedentary parasite of soybean. Its life cycle consists of four juvenile stages and the adult stage (Fig. 1). After hatching, the preinfective, second-stage juveniles (pi-J2s) migrate toward the root and burrow into it. The infective J2s then migrate toward the root stele, using a tubular mouthpiece known as a stylet to slice through cells as it migrates. The nematode selects a pericycle cell or neighboring root cell, for its feeding site. The infective J2s then presumably inject substances through its stylet into the cell that then cause major changes in the physiology of the root cell. The cell walls of the selected cell dissolve, permitting their fusion with neighboring cells. The repeated cell fusion events produce a syncytium that contains approximately 200 merged root cells and serves as the H. glycines nurse cell (Jones, 1981; Jung and Wyss, 1999). After the establishment of the syncytium, male nematodes feed until the end of their J3 stage. Subsequently, the males stop feeding and molt into vermiform J4 males. The males burrow out of the root in preparation for copulation. In contrast, the females remain sedentary after the establishment of their feeding site. The female nematodes increase in size while undergoing both J3 and J4 molts, then mature, becoming adult feeding females. During their growth, the posterior end of the female pushes out of the root boundary. This permits access to the male for copulation outside of the root. Subsequently, the female develops into the hardened cyst that encases the eggs. Cysts, encasing the eggs, are able to remain dormant in the soil for up to 9 years (Inagaki and Tsutsumi, 1971).
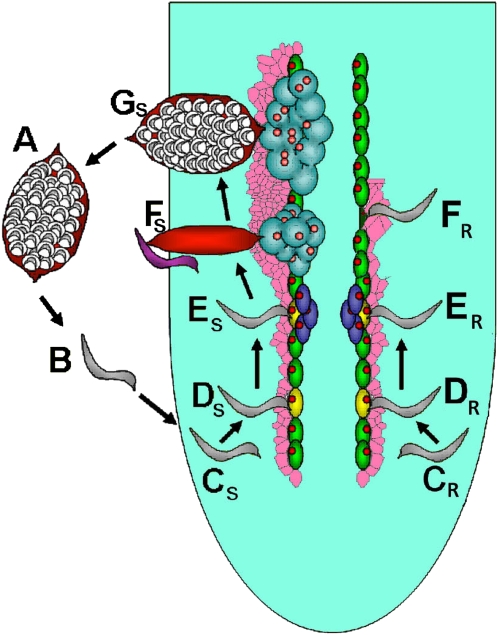
Figure 1.
Life cycle of SCN. A, Cysts. B, pi-J2s (gray) hatch and migrate toward the root of soybean. CS and CR, i-J2 nematodes burrow into the root and migrate toward the pericycle (green). DS and DR, i-J2s select a cell (yellow) for feeding site establishment. ES, i-J2 nematodes have molted into J3. ER, i-J2 nematodes do not increase in size. FS, The J3s undergo a subsequent molt into J4 nematodes. Meanwhile, the female continues to grow circumferentially as it feeds. The male discontinues feeding at the end of its J3 stage. Male and female J4 nematodes become adults. The vermiform male (blue) burrows outside the root and subsequently copulates with the female. FR, The syncytium collapses and the nematodes do not grow. GS, After approximately 30 d, the female with eggs is clearly visible and emerging from the root. Figure adapted from Klink et al. (2009a).
Soybean resistance to SCN is multigenic, composed of both dominant and recessive genes (for review, see Concibido et al., 2004). Over 118 sources of soybean resistance to SCN have been identified, however, only a few of these sources are used for commercial development in the United States (Shannon et al., 2004). The source of most of the resistance that is bred into greater than 95 of the soybean commercial cultivars in the United States is from two genotypes, cv Peking and PI 88788. Anatomical studies have shown that the resistant responses of Peking and PI 88788 are fundamentally different. Peking resistance involves a rapid and potent response at the site of infection while a more delayed response is found in PI 88788 (Luedders and Anand, 1989). The understanding of resistance to SCN has been aided by genetic marker technology and quantitative trait loci (QTL) mapping. A QTL is defined as a phenotypic characteristic that varies in degree. This variance can be attributed to the interactions between two or more genes and their environment. Importantly, QTLs may not necessarily be the genes themselves, but are stretches of DNA that are closely linked to the genes that underlie the trait in question. Those studies have identified QTLs that map to 17 linkage groups. Peking has nine QTLs that map to different linkage groups (for review, see Concibido et al., 2004). PI 88788 has five or fewer resistance QTLs.
GENE EXPRESSION IN SOYBEAN ROOTS DURING SCN INVASION
The identification of the genes involved in the resistant response has been hampered by the complex nature of the soybean genome. However, methods that study gene expression during a process are a useful way to correlate genes with a particular process. Microarrays offer a means to investigate the activity of all genes within a genome. Microarray analysis has been applied to the understanding of plant pathogenic nematode infection. Puthoff et al. (2003) studied Arabidopsis (Arabidopsis thaliana) infection with sugar beet (Beta vulgaris) cyst nematode and SCN using commercially available microarrays, resulting in the identification of genes whose expression changes during infection. However, initially exploiting this technology for agriculturally relevant plants such as soybean was limited by the lack of commercially available microarrays. Microarray analyses using SCN-infected soybean roots was conducted using microarrays constructed in the laboratory using inserts from complementary DNA (cDNA) libraries (Alkharouf et al., 2004). From those noncommercially available microarrays, transcriptional changes were identified that occur during infection in whole soybean roots during infection by SCN (Khan et al., 2004; Alkharouf et al., 2006). While array hybridization technology improved, the major obstacle to optimizing the value of the data was the process of gene annotation. Those studies involved computational methods that were housed with the large amount of microarray data (i.e. the Soybean Genomics and Microarray Database for public use). The Soybean Genomics and Microarray Database provided online analytical processing of the data, so no outside software was needed to mine the data (Alkharouf and Matthews, 2004; Alkharouf et al., 2005).
Early attempts at understanding nematode infection of plants were limited to the period when the nematodes have selected their feeding sites (Puthoff et al., 2003; Khan et al., 2004). However, the infection of plants by parasitic nematodes is composed of a period before the selection of feeding sites and a period after which the nematodes select cells that develop into nurse cells. In soybean, the time prior to feeding cell selection is the first 24 h of infection and nothing was known of how plants respond to the presence of the nematode within its tissues prior to feeding site selection. The first time course microarray analysis of the soybean-SCN system studied seven time points to investigate the soybean-SCN interaction, including time points both prior to (6 and 12 h postinoculation [hpi]) and after feeding site selection (1, 2, 4, 6, and 8 d postinoculation [dpi]; Alkharouf et al., 2006). Numerous genes were identified with induced levels of gene expression before feeding site selection during a susceptible reaction. These genes included the pathogenesis-related genes SAM22 (PR-10) and Kunitz trypsin inhibitor. Other genes included those with antimicrobial activity such as germin-like protein, several unknown stress-related genes, and a peroxidase precursor. Other genes included phospholipase D, 12-oxyphytodienoate reductase, genes in metabolism such as a trehalose-6-P synthase homolog, and genes involved in secondary metabolism such as coumarate CoA ligase, and components of the phenylpropanod pathway such as deoxychalcone synthase and chalcone reductase. While the analyses studied probe sets representing approximately 6,000 cDNA clones, the work confirmed other microarray and gene expression analyses performed by other labs. With the availability of the commercial Affymetrix soybean GeneChip containing 37,744 soybean transcripts (35,611 transcripts) and 7,539 SCN probe sets for 7,431 transcripts, the expression of a broader spectrum of genes could be monitored. Ithal et al. (2007) studied three time points after feeding site selection occurred, 2, 5, and 10 dpi. They noted an increase in expression of genes involved in lignin and flavonoid biosynthesis, phenolic compounds, cell wall modification, defense, and stress. Among these were genes related to PR-5, PR-1a, and expansin. The major focus of their work was to determine the expression of a panel of putative parasitism genes of SCN that were identified in a series of previous experiments (Gao et al., 2001, 2003). The analysis determined that numerous parasitism genes were induced during the parasitic stages of infection.
The soybean-SCN system is an exceptional model to compare gene expression during infection, because both resistant and susceptible reactions can be obtained and studied in the same genotype (Klink et al., 2007b, 2007c, 2009a, 2009b). A further advantage of the soybean-SCN model system is the availability of well-defined SCN populations (for review, see Niblack and Riggs, 2004). Soybean can be resistant to one SCN population and susceptible to another; likewise, the nematode may be successful infecting one soybean genotype, yet cannot develop to maturity in another genotype. The HG types for NL1-RHg and TN8 populations used by the Matthews laboratory in studies discussed below were HG type 7 and HG type 1.3.6.7, respectively, as determined independently in the laboratory of Dr. Terry Niblack (Department of Crop Sciences, University of Illinois) in 2007 according to the HG type test procedures of Niblack et al. (2002). The susceptible reaction was obtained by using TN8 (Niblack et al., 2002), while the resistant reaction was obtained using NL1-RHg. Klink et al. (2007b) examined gene expression at 12 hpi, 3 and 8 dpi in both a resistant and susceptible reaction using whole infected roots from Peking infected with incompatible (NL1-RHg; HG type 7) and compatible (TN8; HG type 1.3.6.7) populations of SCN (Klink et al., 2007b). Those analyses have shown that soybean can differentiate between the different populations of nematodes as early as 12 hpi (Klink et al., 2007b), which is before the nematode has selected its feeding site. The analyses also demonstrated that there are differences in gene expression in the two SCN populations at 12 hpi.
Recently, two-dimensional electrophoresis was used to explore resistance and susceptibility of soybean to SCN (Afzal et al., 2009). More than 1,000 protein spots were resolved using extracts from near-isogenic lines of soybean containing or lacking the rhg1 locus, a major component of SCN resistance, 10 dpi and without SCN infection. Pathway alterations included those involved in phytoalexin and inositol production and glycolysis.
SCN TRANSCRIPTOME ANALYSIS
Changes in gene expression occur during the life cycle of SCN (Ithal et al., 2007; Klink et al., 2007b, 2009a, 2009c). Those differences occur abruptly during the transition of the nematode from a mobile J2 to a sedentary feeding parasite (Klink et al., 2007b, 2009a, 2009c). Numerous analyses have demonstrated changes in SCN gene expression at various stages of its development. Some studies have focused on the dorsal and esophageal glands that are the sites of synthesis of substances that facilitate parasitism. Importantly, a panel of putative parasitism genes was identified through the creation and analysis of a gland cell cDNA library (Gao et al., 2001, 2003). Some of the cDNAs were related to enzymes involved in cell wall degradation and had signal peptides homologous to those involved in secretion. Prior in situ hybridization experiments had already confirmed that they localized to the esophageal glands (Gao et al., 2001, 2003). Transcriptomic analyses of putative SCN parasitism genes determined that they were expressed during the parasitism stages of infection during a susceptible reaction (Ithal et al., 2007). The study used infective J2s at 2 dpi, J3s at 5 dpi, and maturing males and females at 10 dpi. These analyses were followed up by a complete time course analysis of all stages of SCN development during a compatible interaction (Elling et al., 2009). The aforementioned experiments determined gene expression as it pertained to a compatible reaction. However, due to the design of the experiments, they could not differentiate what genes were essential for parasitism. This is because experiments of nematodes undergoing an incompatible reaction were not investigated. Cytological and ultrastructural observations have shown that the early stages of an incompatible and compatible interaction, between 1 and 4 dpi, appear to be the same (Endo, 1965; Riggs et al., 1973; Kim et al., 1987, 1992; Klink et al., 2009a, 2009b). However, between 4 and 5 dpi, the incompatible reaction becomes evident as syncytia collapse and nematodes, depending on the soybean genotype, fail to grow.
Subsequent, comparative analyses investigating H. glycines gene expression demonstrated that different races of SCN that elicit a resistant or susceptible reaction in soybean have different transcriptional profiles at the pi-J2 stage even before they infect roots. Microarray analyses were performed on soybean cv Peking infected with the incompatible SCN population NL1-RHg, HG type 7, that has been genetically inbred and used for decades for SCN research. The compatible population was the genetically inbred TN8 (Niblack et al., 2002). An expression analysis determined that 71 genes were induced in the incompatible NL1-RHg population as compared directly to the compatible TN8 (baseline) at the pi-J2 stage (Klink et al., 2009a). Of those, 19 genes were induced 5-fold or greater. Those genes included two G23G12 putative gland proteins and two Hgg-20 genes and an unknown homolog of Caenorhabditis elegans temporarily assigned gene name 287 (Klink et al., 2009a). There were also 44 suppressed genes in NL1-RHg as compared to TN8 (Klink et al., 2009a). Genes suppressed more than 5-fold included several esophageal gland proteins. These results meant that there were significant transcriptomic differences present between the two populations even before the nematodes had infected the plant tissue. Subsequent experiments at the 12 hpi and 3 dpi time points demonstrated fewer differences in gene expression. Importantly, there were nine induced and 10 suppressed genes identified at 3 dpi (Klink et al., 2009a). This is the time when incompatible NL1-RHg and compatible TN8 populations are establishing feeding sites. Therefore, few obvious differences in gene expression are present between the two nematode populations at the 3 dpi time point. This is important to note because the early responses of the syncytium during the resistant and susceptible reactions appear the same both cytologically and ultrastructurally (Endo, 1965; Riggs et al., 1973; Acido et al., 1984; Kim et al., 1987; Klink et al., 2007a, 2007b). However, by 8 dpi, NL1-RHg had 13 induced and 1,668 suppressed genes (Klink et al., 2009a). The experiments possibly identified many genes essential for parasitism. The most highly suppressed genes of known function were a steroid α-reductase and a Ser protease that are important for nematode nutrition.
GENE EXPRESSION IN THE SYNCYTIUM
Changes in gene expression in soybean roots exhibiting the incompatible and compatible reactions may not fully reflect what gene expression changes occur at the feeding site (syncytium), wherein the many changes in cellular morphology occur as described previously. Understanding the gene expression pattern of the resistance reaction locally at the nematode feeding site may lead to an understanding of the genetic program underlying the response. However, studying gene expression specifically at the feeding site has been complicated because the feeding sites are difficult to isolate and by technical limitations imposed by the quantity of material that can be faithfully obtained. LCM bypasses these problems and provides a high degree of precision and accuracy to isolate syncytial cells (Fig. 2; Klink et al., 2005, 2007b, 2009b). LCM and microarrays were used to investigate the syncytium during soybean infection by SCN during a susceptible (Klink et al., 2005) and resistant reactions (Klink et al., 2007b, 2009b). In each of those analyses the identification of syncytia was aided by using the nematode as an in situ physical marker for the syncytium. These studies encompassed the construction of cDNA libraries, the cloning of full-length cDNAs, the generation of markers for RNA in situ hybridization experiments, the identification of probes for protein expression experiments, and a quantitative analysis of gene expression (Klink et al., 2005).
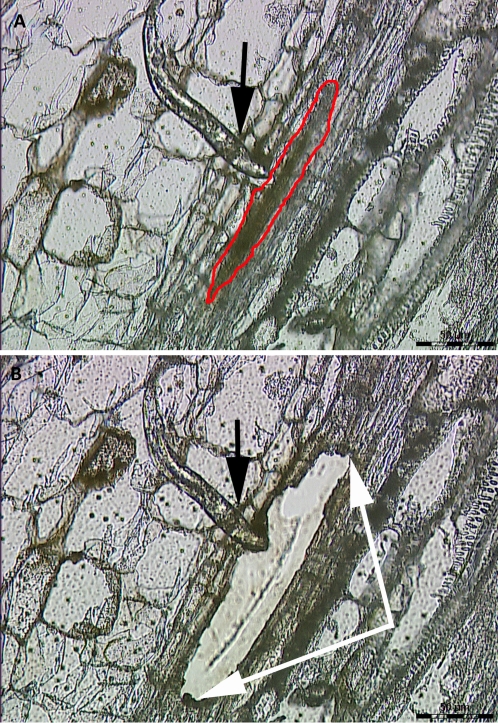
Figure 2.
A microdissected syncytium undergoing a resistant reaction. A, Before LCM. B, After LCM. Red line, perimeter of the syncytium; black arrow, head of nematode; white arrows, boundary of the microdissected syncytium. Figure adapted from Klink et al. (2009b).
The first comparative microarray expression analysis of LCM isolated syncytia undergoing resistant or susceptible reactions revealed distinct differences in gene expression between syncytia undergoing resistant and susceptible responses (Klink et al., 2007b). The analysis identified 136 genes that were induced specifically in syncytia undergoing a resistant reaction. This early stage was characterized by induced levels of genes encoding lipoxygenase (LOX), heat shock protein 70, superoxide dismutase, an arabinogalactan, a mitotic checkpoint protein, an expansin, a WRKY transcription factor, a calreticulin, and others (Klink et al., 2007b). Several of these genes pertain to important modes of plant defense. Of note, LOX is associated with plant development, responses to environmental change, and challenge with pathogens (Song et al., 1993). LOXs are non-heme dioxygenases, catalyzing the oxygenation of polyunsaturated fatty acids. Ultimately, this activity leads to the generation of hydroperoxides that can be converted into (1) stable aldehydes, (2) hydroxy and epoxy fatty acids that exhibit antimicrobial activity, and (3) jasmonic acid. Jasmonic acid is derived from the LOX product 13-hydroperoxy-octadecatrienoic acid. In addition to the role of jasmonic acid in resistance to SCN in the syncytia, they noted an increase in expression of numerous genes encoding enzymes involved in cell wall modification. It has been reported that LOX is involved in resistance to Meloidogyne incognita (Gao et al., 2008). In contrast, 163 genes were suppressed specifically in syncytia undergoing a resistant reaction (Klink et al., 2007b). Ithal et al. (2007) also examined gene expression in syncytia during the susceptible reaction and also concluded that the jasmonic acid biosynthetic pathway appears to be down-regulated, while genes encoding proteins that modify the cell wall and regulate lignifications are up-regulated. Therefore, the earlier stage of resistance includes gene expression that is specific to syncytia undergoing resistant and susceptible reactions. A second phase of gene expression clearly differentiates resistant from susceptible syncytia between the 3 and 8 dpi time points.
NEW MODES OF RESISTANCE
Ultimately, the goal of this research is to develop new modes of resistance to nematodes to improve crop yield. Transgenic plants that overexpress soybean genes correlated with resistance or that silence soybean genes important to syncytium development and nematode success are two obvious areas to explore. However, resistance and susceptibility are complex and a mixture of several genes overexpressed and silenced may be required. Also showing promise is the approach of silencing of nematode genes by producing RNAi at the feeding site in the soybean root.
Successful silencing of SCN gene expression using RNAi was first demonstrated in SCN (Urwin et al., 2002). In those experiments, double-stranded RNA was synthesized in vitro and SCN was soaked for a period of time. Subsequently the nematodes were allowed to infect soybean; their fecundity was decreased (Urwin et al., 2002). Steeves et al. (2006) demonstrated that transgenic soybean expression of an RNAi-inducing construct for the major sperm protein of SCN reduced egg production up to 68%. Alkharouf et al. (2007) performed double-stranded RNA soaking experiments of genes that were identified to be conserved to C. elegans genes with lethal mutant or RNAi phenocopies, indicating that a high proportion of nematodes could be killed using RNAi. In related experiments, Klink et al. (2009c) functionally tested putative parasitism genes that were identified by microarray analyses. Screening of Affymetrix microarrays resulted in the identification of 229 highly conserved genes. Of those, 131 also had homologs with lethal RNAi phenocopies in C. elegans. Of those, 32 were induced during the parasitic stages of SCN development (Klink et al., 2009c). Four genes were selected for their expression in in planta experiments as performed by Huang et al. (2006) and Steeves et al. (2006). The development of female nematodes was reduced by 80% or more (Klink et al., 2009c). This result demonstrated that transcriptomic analyses could identify genes useful to retard or stop the development of female nematodes.
CONCLUSION
With the sequencing of the soybean and SCN genomes and development of microarrays with many soybean and SCN genes represented, major advances in understanding the interactions of soybean with SCN have occurred. In addition, practical application of this knowledge is on the near horizon. Continued work in this area combined with technology advancements such as deep sequencing of RNA transcripts and improved genome annotation promise to provide a basic understanding of soybean interactions with nematodes and real solutions to real problems.
Acknowledgments
Dr. Gary Lawrence, Department of Entomology and Plant Pathology, Mississippi State University, provided helpful insight. Mention of trade names or commercial products in this article is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the U.S. Department of Agriculture.
This work was supported by the United Soybean Board (grant no. Y9254 to B.F.M.), and by the Research Initiation Program Grant at Mississippi State University and the Mississippi Soybean Promotion Board (to V.P.K.).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Vincent P. Klink (vklink@biology.msstate.edu).
References
- Abad P, Gouzy J, Aury JM, Castagnone-Sereno P, Danchin EG, Deleury E, Perfus-Barbeoch L, Anthouard V, Artiguenave F, Blok VC, et al (2008) Genome sequence of the metazoan plant-parasitic nematode Meloidogyne incognita. Nat Biotechnol 26 909–915 [DOI] [PubMed] [Google Scholar]
- Acido JR, Dropkin VH, Luedders VD (1984) Nematode population attrition and histopathology of Heterodera glycines-soybean associations. J Nematol 16 48–57 [PMC free article] [PubMed] [Google Scholar]
- Afzal AJ, Natarajan A, Saini N, Iqbal MJ, Geisler M, El Shemy HA, Mungur R, Willmitzer L, Lightfoot DA (2009) The nematode resistance allele at the rhg1 locus alters the proteome and primary metabolism of soybean roots. Plant Physiol 151 1264–1280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alkharouf N, Khan R, Matthews BF (2004) Analysis of expressed sequence tags from roots of resistant soybean infected by the soybean cyst nematode. Genome 47 380–388 [DOI] [PubMed] [Google Scholar]
- Alkharouf N, Klink VP, Matthews BF (2007) Identification of Heterodera glycines (soybean cyst nematode [SCN]) DNA sequences with high similarity to those of Caenorhabditis elegans having lethal mutant or RNAi phenotypes. Exp Parasitol 115 247–258 [DOI] [PubMed] [Google Scholar]
- Alkharouf N, Matthews BF (2004) SGMD: the soybean genomics and microarray database. Nucleic Acids Res 32 D398–D400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alkharouf NW, Jamison DC, Matthews BF (2005) Online analytical processing (OLAP): a fast and effective data mining tool for gene expression databases. J Biomed Biotechnol 2 181–188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alkharouf NW, Klink VP, Chouikha IB, Beard HS, MacDonald MH, Meyer S, Knap HT, Khan R, Matthews BF (2006) Timecourse microarray analyses reveals global changes in gene expression of susceptible Glycine max (soybean) roots during infection by Heterodera glycines (soybean cyst nematode). Planta 224 838–852 [DOI] [PubMed] [Google Scholar]
- Concibido VC, Diers BW, Arelli PR (2004) A decade of QTL mapping for cyst nematode resistance in soybean. Crop Sci 44 1121–1131 [Google Scholar]
- Elling AA, Mitreva M, Gai X, Martin J, Recknor J, Davis EL, Hussey RS, Nettleton D, McCarter JP, Baum TJ (2009) Sequence mining and transcript profiling to explore cyst nematode parasitism. BMC Genomics 10 58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endo BY (1965) Histological responses of resistant and susceptible soybean varieties, and backcross progeny to entry development of Heterodera glycines. Phytopathology 55 375–381 [Google Scholar]
- Gao B, Allen R, Maier T, Davis EL, Baum TJ, Hussey RS (2001) Identification of putative parasitism genes expressed in the esophageal gland cells of the soybean cyst nematode Heterodera glycines. Mol Plant Microbe Interact 14 1247–1254 [DOI] [PubMed] [Google Scholar]
- Gao B, Allen R, Maier T, Davis EL, Baum TJ, Hussey RS (2003) The parasitome of the phytonematode Heterodera glycines. Mol Plant Microbe Interact 16 720–726 [DOI] [PubMed] [Google Scholar]
- Gao X, Starr J, Göbel C, Engelberth J, Feussner I, Tumlinson J, Kolomiets M (2008) Maize 9-lipoxygenase ZmLOX3 controls development, root-specific expression of defense genes, and resistance to root-knot nematodes. Mol Plant Microbe Interact 21 98–109 [DOI] [PubMed] [Google Scholar]
- Huang G, Allen R, Davis EL, Baum TJ, Hussey RS (2006) Engineering broad root-knot resistance in transgenic plants by RNAi silencing of a conserved and essential root-knot nematode parasitism gene. Proc Natl Acad Sci USA 103 14302–14306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inagaki H, Tsutsumi M (1971) Survival of the soybean cyst nematode, Heterodera glycines Ichinohe (Tylenchida: Heteroderidae) under certain storage conditions. Appl Entomol Zool (Jpn) 8 53–63 [Google Scholar]
- Ithal N, Recknor J, Nettleston D, Hearne L, Maier T, Baum TJ, Mitchum MG (2007) Parallel genome-wide expression profiling of host and pathogen during soybean cyst nematode infection of soybean. Mol Plant Microbe Interact 20 293–305 [DOI] [PubMed] [Google Scholar]
- Jones MGK (1981) The development and function of plant cells modified by endoparasitic nematodes. In BM Zuckerman, RA Rohde, eds, Plant Parasitic Nematodes, Vol III. Academic Press, New York, pp 255–279
- Jung C, Wyss U (1999) New approaches to control plant parasitic nematodes. Appl Microbiol Biotechnol 51 439–446 [Google Scholar]
- Khan R, Alkharouf N, Beard HS, MacDonald M, Chouikha I, Meyer S, Grefenstette J, Knap H, Matthews BF (2004) Resistance mechanisms in soybean: gene expression profile at an early stage of soybean cyst nematode invasion. J Nematol 36 241–248 [PMC free article] [PubMed] [Google Scholar]
- Kim YH, Riggs RD, Kim KS (1987) Structural changes associated with resistance of soybean to Heterodera glycines. J Nematol 19 177–187 [PMC free article] [PubMed] [Google Scholar]
- Klink VP, Hosseini P, MacDonald MH, Alkharouf N, Matthews BF (2009. a) Population-specific gene expression in the plant pathogenic nematode Heterodera glycines exists prior to infection and during the onset of a resistant or susceptible reaction in the roots of the Glycine max genotype Peking. BMC Genomics 10 111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klink VP, Hosseini P, Matsye P, Alkharouf N, Matthews BF (2009. b) A gene expression analysis of syncytia laser microdissected from the roots of the Glycine max (soybean) genotype PI 548402 (Peking) undergoing a resistant reaction after infection by Heterodera glycines (soybean cyst nematode). Plant Mol Biol (in press) [DOI] [PubMed]
- Klink VP, Kim KH, Martins VE, MacDonald MH, Beard HS, Alkharouf NW, Park SC, Matthews BF (2009. c) A correlation between host-mediated expression of parasite genes as tandem inverted repeats and abrogation of the formation of female Heterodera glycines cysts during infection of Glycine max. Planta 230 53–71 [DOI] [PubMed] [Google Scholar]
- Klink VP, MacDonald M, Alkharouf N, Matthews BF (2005) Laser capture microdissection (LCM) and expression analyses of Glycine max (soybean) syncytium containing root regions formed by the plant pathogen Heterodera glycines (soybean cyst nematode). Plant Mol Biol 59 969–983 [DOI] [PubMed] [Google Scholar]
- Klink VP, Martins VE, Overall CC, Alkharouf N, MacDonald MH, Matthews BF (2007. a) A decline in transcript abundance for Heterodera glycines homologs of Caenorhabditis elegans uncoordinated genes accompanies its sedentary parasitic phase. BMC Dev Biol 7 35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klink VP, Overall CC, Alkharouf N, MacDonald MH, Matthews BF (2007. b) Laser capture microdissection (LCM) and comparative microarray expression analysis of syncytial cells isolated from incompatible and compatible soybean roots infected by soybean cyst nematode (Heterodera glycines). Planta 226 1389–1409 [DOI] [PubMed] [Google Scholar]
- Klink VP, Overall CC, Alkharouf N, MacDonald MH, Matthews BF (2007. c) A comparative microarray analysis of an incompatible and compatible disease response by soybean (Glycine max) to soybean cyst nematode (Heterodera glycines) infection. Planta 226 1423–1447 [DOI] [PubMed] [Google Scholar]
- Luedders VD, Anand SC (1989) Attempt to select a cyst nematode population on soybean plant introduction 437654. J Nematol 21: 264–267 [PMC free article] [PubMed]
- Niblack TL, Arelli PR, Noel GR, Opperman CH, Orf JH, Schmitt DP, Shannon JG, Tylka GL (2002) A revised classification scheme for genetically diverse populations of Heterodera glycines. J Nematol 34 279–288 [PMC free article] [PubMed] [Google Scholar]
- Niblack TL, Riggs RD (2004) Variation in virulence phenotypes. In DP Schmitt, JA Wrather, RD Riggs, eds, Biology and Management of Soybean Cyst Nematode, Ed 2. Schmitt & Associates of Marceline, Marceline, MO, pp 57–71
- Puthoff DP, Nettleton D, Rodermel SR, Baum TJ (2003) Arabidopsis gene expression changes during cyst nematode parasitism revealed by statistical analyses of microarray expression profiles. Plant J 33 911–921 [DOI] [PubMed] [Google Scholar]
- Riggs RD, Kim KS, Gipson I (1973) Ultrastructural changes in Peking soybeans infected with Heterodera glycines. Phytopathology 63 76–84 [Google Scholar]
- Shannon JG, Arelli PR, Young LD (2004) Breeding for resistance and tolerance. In DP Schmitt, JA Wrather, RD Riggs, eds, Biology and Management of Soybean Cyst Nematode, Ed 2. Schmitt & Associates of Marceline, Marceline, MO, pp 155–180
- Sinclair JB, Backman PA (1989) Compendium of Soybean Diseases. American Phytopathological Society, St. Paul
- Song WC, Baertschi SW, Boeglin WE, Harris TM, Brash AR (1993) Formation of epoxyalcohols by a purified allene oxide synthase: implications for the mechanism of allene oxide synthesis. J Biol Chem 268 6293–6298 [PubMed] [Google Scholar]
- Steeves RM, Todd TC, Essig JS, Trick HN (2006) Transgenic soybeans expressing siRNAs specific to a major sperm protein gene suppress Heterodera glycines reproduction. Funct Plant Biol 33 991–999 [DOI] [PubMed] [Google Scholar]
- Urwin PE, Lilley CJ, Atkinson HJ (2002) Ingestion of doublestranded RNA by preparasitic juvenile cyst nematodes leads to RNA interference. Mol Plant Microbe Interact 15 747–752 [DOI] [PubMed] [Google Scholar]