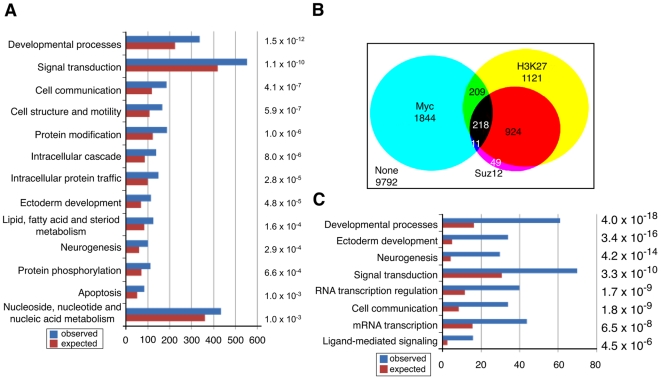
Figure 3. c-Myc, and Polycomb proteins share target genes.
(A) Gene Ontology categories of c-Myc associated genes in murine ES cells. Functional categories derived for genes enriched at least 4-fold for c-Myc binding and possessing an ACME p<0.0001 in the ChIP-chip assay (see Materials and Methods and Supplementary Table S1). The numbers to the right represent the p-values for the statistical over-representation in each category. (B) Venn diagram depicting the overlap among murine ES cell genes bound by c-Myc (this paper), Suz12, and those displaying trimethylation of H3-K27 [16]. Nearly 96% of promoters bound by Suz12 also show the H3-K27me3 mark. Of genes associated with Myc (2282) 17% are associated with the H3-K27me3 mark and 9.5% associated with both Suz12 and H3-K27me3 (for gene list see Supplementary Table S3). (C) Gene ontology categories of gene displaying overlap among c-Myc, Suz12 binding and H3-K27me3 marks. Functional categories were derived for genes enriched at least 4-fold for c-Myc binding in the ChIP-chip assay (see Methods and Supplementary Table S1). Suz12 and H3-K27me3 bound genes were previously described [16]. Numbers to the right are p-values for the statistical over-representation among genes within a functional category.