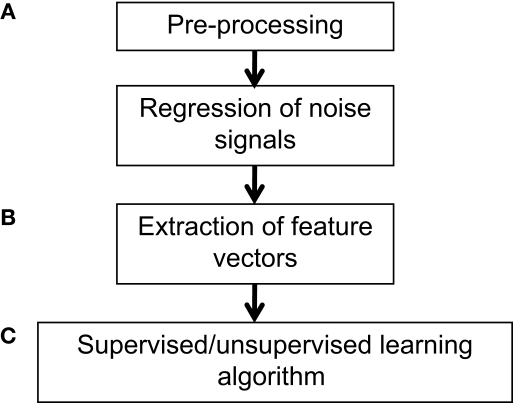
Figure 1.
Example workflow for pattern classification analysis. (A) Preprocessing of fMRI data can follow that used for conventional GLM analyses, consisting of slice-timing correction, realignment, and optional spatial normalization and smoothing steps. However, while the GLM can reduce potential noise sources by including signals such as motion parameters and physiological noise models as covariates in the GLM design matrix, MVPA does not typically have a framework for modeling confounds. Thus, when applying MVPA to the raw time-series, expected confounds should be removed from the data prior to performing MVPA, such as by obtaining the residuals from a regression over the nuisance variables. In addition, if the features are activation patterns (parameter estimates for a particular model), one may wish to first run a GLM analysis on the fMRI data and extract the relevant contrast estimates. (B) Next, the pre-processed data should be transformed into “feature vectors”. This involves creating, for each data point (e.g., subject) a vector in which the ith entry corresponds to the value of the ith feature for that data point. Feature selection may be applied to reduce the number of entries in the feature vectors. (C) At this stage, one may choose to run an unsupervised learning algorithm to characterize patterns in the data, or to run a supervised learning algorithm in conjunction with cross-validation. The parameters of the trained model can be examined for further insight, and the model may also be applied to make predictions on additional datasets so as to further explore its generalization ability.