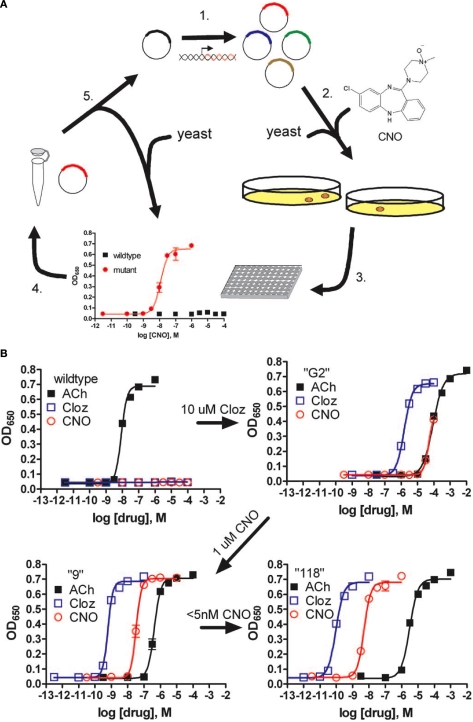
Figure 1.
Pharmacological profiles of an rM3Δi3 receptor mutant selected during directed molecular evolution for CNO responsiveness. (A) Experimental design for directed evolution of mammalian GPCRs in yeast to create DREADDs. (1) Libraries of randomly mutated rM3Δi3 receptors were produced by mutagenic PCR; (2) yeast-expressing mutant receptors activated by synthetic ligands (e.g., CNO) were selected for by growth on nutrient deficient medium; (3) mutants were verified by secondary liquid growth assays in 96-well plates; (4) plasmid DNA was isolated from yeast; (5) clones were retransformed into yeast to pharmacologically profile mutants by liquid growth assays, and those with desirable properties were sequenced and remutagenized for subsequent rounds of selection to yield receptors with higher potency. (B) Optical density at 650 nm of liquid cultures of yeast transformed with either wild type, clone “G2” (first library, 10 μM clozapine screen), clone “9” (second library, 1 μM CNO screen), or clone “118” (third library, 5-nM CNO screen) rM3Δi3 receptors incubated with ACh (■), clozapine (□), or CNO (E). Data shown are mean ± SEM values of a representative experiment performed with two independent yeast transformants grown for each clone. Figure used with permission: Copyright (2007) National Academy of Sciences, USA Armbruster et al. (2007).