Abstract
Influenza A virus M2 (A/M2) and the influenza B virus BM2 are both small integral membrane proteins that form proton-selective ion channels. Influenza A virus A/M2 channel is the target of the antiviral drug amantadine (and its methyl derivative rimantadine), whereas BM2 channel activity is not affected by the drug. The atomic structure of the pore–transmembrane (TM) domain peptide has been determined by x-ray crystallography [Stouffer et al. (2008) Nature 451:596–599] and of a larger M2 peptide by NMR methods [Schnell and Chou (2008) Nature 451:591–595]. The crystallographic data show electron density (at 3.5 Å resolution) in the channel pore, consistent with amantadine blocking the pore of the channel. In contrast, the NMR data show 4 rimantadine molecules bound on the outside of the helices toward the cytoplasmic side of the membrane. Drug binding includes interactions with residues 40–45 and a polar hydrogen bond between rimantadine and aspartic acid residue 44 (D44). These 2 distinct drug-binding sites led to 2 incompatible drug inhibition mechanisms. We have generated chimeric channels between amantadine-sensitive A/M2 and amantadine-insensitive BM2 designed to define the drug-binding site. Two chimeras containing 5 residues of the A/M2 ectodomain and residues 24–36 of the A/M2 TM domain show 85% amantadine/rimantadine sensitivity and specific activity comparable to that of WT BM2. These functional data suggest that the amantadine/rimantadine binding site identified on the outside of the 4 helices is not the primary site associated with the pharmacologic inhibition of the A/M2 ion channel.
Keywords: inhibition, binding site, proton-selective
Influenza A virus M2 protein (A/M2) and the influenza B virus BM2 protein are both small integral membrane proteins of 97 and 108 aa residues, respectively, and they both adopt an NoutCin orientation (1, 2). The A/M2 protein has a 24-residue N-terminal extracellular domain, a single internal hydrophobic domain of 19 residues that acts as a transmembrane (TM) domain and forms the pore of the channel, and a 54-residue cytoplasmic tail (1). The BM2 protein consists of a 7-residue ectodomain, a 19-residue TM domain, and an 82-residue cytoplasmic tail (2). Both A/M2 and BM2 are proton-selective ion channels (3–5). The functionally active A/M2 and BM2 channel are homotetramers (2, 6, 7), and both A/M2 and BM2 have a centrally located pore for proton conduction (8–13). However, the only amino acid identity between the 2 channels is the HXXXW motif of the inner membrane-spanning amino residues (residues 37–41 in A/M2 and residues 19–23 in BM2, respectively). The high proton selectivity of A/M2 is conferred by histidine residue 37 (14–16), and the channel gate is conferred by tryptophan 41 (17). In the BM2 channel His 19 and Trp 23 are thought to play similar roles (5).
Influenza A virus A/M2 channel is the target of the antiviral drug amantadine (and its methyl derivative rimantadine) (18; reviewed in refs. 19 and 20), whereas neither influenza B virus growth (21) nor the BM2 channel activity are affected by the drugs (5). Influenza A viruses that mutate and become drug resistant contain mutations in the A/M2 TM domain, and naturally arising mutations occur predominantly at TM domain residue Ser 31 but also to a lesser extent at Leu 26, Val 27, Ala 30, and Gly 34 (22).
The atomic structure of the A/M2 TM domain has been determined using x-ray crystallography (23), and the atomic structure of a larger peptide (A/M2 residues 18–60) has been determined using NMR methods (24). As predicted from earlier biochemical and NMR studies (9, 25–29), the A/M2 TM domain forms a 4-helix bundle, with His 37 forming the pH sensor and Trp 41 indole rings, which are within van der Waal's distance from each other, forming the channel gate. The Trp-41 indole rings, in the channel closed state, would prohibit the passage of water or ions, and the gate is further stabilized by intersubunit hydrogen bonds with Asp-44.
It has long been thought that amantadine must bind to the A/M2 channel in the region where mutations arise that lead to amantadine resistance, residues 27–34 (22; reviewed in ref. 30). The x-ray structure of the TM domain, in the presence of amantadine but not in its absence, shows electron density in the channel pore between residues 27 and 34, consistent in dimensions with it representing a single molecule of the drug (23). However, the atomic resolution (3.5 Å) does not permit the definitive assignment of the density to amantadine. In contrast, Schnell and Chou (24) did not find amantadine in the channel pore but found 4 molecules of rimantadine bound on the outside of the 4-helix bundle facing the lipid bilayers with interactions with A/M2 TM domain residues 40–45. A hydrogen bond between rimantadine and aspartic acid residue 44 (D44) was thought to be particularly important (24).
Two models for amantadine inhibition have arisen from the structural data. Stouffer et al. (23) proposed that amantadine physically blocked the ion conductance pathway by filling the restricted vestibule between residues V27 and G34. The rate of channel block is 105-fold slower than calculated for a channel with a wide opening (31), and the restricted vestibule may provide an explanation for the slow kinetics of entry of the drug. In this model the drug presumably enters the vestibule by means of rare conformational changes or laterally from the bilayer phase. This physical block model is consistent with the observation that when amantadine was injected into the cytoplasm of oocytes there was no inhibition of A/M2 channel activity (32), demonstrating that the pharmacologically relevant binding site is inaccessible to the medium bathing the C-terminal of the protein. Schnell and Chou (24) proposed that the drug inhibited the channel through an allosteric inhibition mechanism, with drug binding stabilizing the closed conformation of the channel. An argument used in support of the long-range effects of the drug binding hypothesis is that the mutation L38F causes amantadine resistance when introduced into the Rostock strain (24, 31, 33). However, we have shown that A/M2 L38F in the A/Udorn/72 genetic background has an amantadine-sensitive channel activity (34). Furthermore, influenza A virus bearing A/M2 L38F is a viable virus sensitive to amantadine (34). Thus, mutations leading to amantadine resistance are found only in the vestibule of the channel. Although the 2 models for drug binding are incompatible, both models do agree with the fact that drug binding is amino acid sequence specific.
The Schnell and Chou model for amantadine binding proposes an important interaction of the drug with D44. However, when we made the D44A mutation and measured the A/M2 D44A channel activity in oocytes of Xenopus laevis, the D44A mutant channel was found to be sensitive to amantadine (34). Furthermore, influenza A virus bearing the A/M2 D44A mutation was found to be a viable virus that is amantadine sensitive and with a growth curve only slightly slower than WT virus (34). Complicating the matter, when Pielak et al. (33) incorporated a peptide containing the D44A mutation into liposomes, they observed a channel activity that was reduced in conductance per tetramer and was amantadine resistant. We do not know the basis for this major discrepancy, but we do note that the liposome assay used was not identical to that used previously (35), and the level of leak current was not evaluated.
In a study of critical residues in A/M2 required for amantadine sensitivity, we constructed a chimeric channel between BM2 (amantadine insensitive) and A/M2 (amantadine sensitive), replacing BM2 residues 6–18 with residues 26–34 from A/M2 (34). When the chimeric channel [(24–36aa A/M2)BM2] was expressed in oocytes of Xenopus laevis it was found that 50% of the channel activity was inhibited by addition of amantadine. However, the activity of this chimeric channel was low as compared with WT BM2 (34). Here we describe greatly improved new chimeric channels that have >85% amantadine/rimantadine sensitivity and specific activity comparable to that of WT BM2. The data have important implications for the mechanism of drug binding.
Results
Chimeric BM2 (19–36aaA/M2) and BM2 d2–5 (19–36aaA/M2) Ion Channels Displayed A/M2-like Ion Channel Properties.
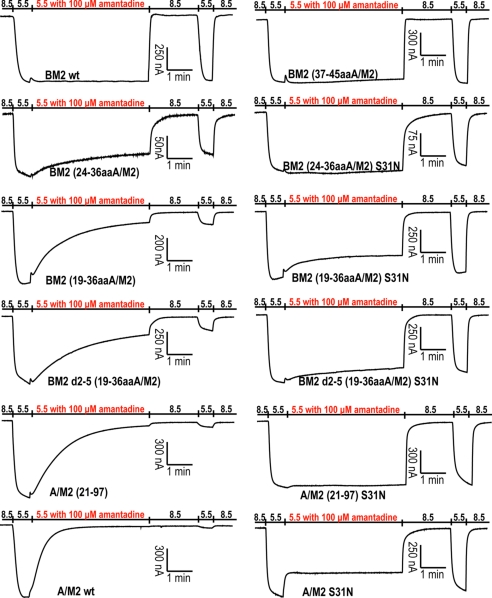
Replacement of the residues of the outer half of the BM2 TM domain (residues 6–18) with the corresponding residues from the A/M2 TM domain (residues 24–36) rendered the resulting chimeric BM2 ion channel [BM2 (24–36aaA/M2)] partially sensitive (≈50%) to amantadine, with a slower onset of inhibition than for WT A/M2 (34). Thus, 13 residues from A/M2 are capable of greatly, but not fully, modifying this important property of the BM2 ion channel. We explored the possibility that the residues distal to these TM domain residues might be important for more complete inhibition by amantadine by testing 2 new constructs, BM2 (19–36aaA/M2) and BM2 d2–5 (19–36aaA/M2), in which the 5 residues of the ectodomain closest to the A/M2 TM domain were transferred to the BM2 protein. In WT M2, cysteine residue 19 forms a disulfide bond (6) (Fig. 1), although the absence of disulfide bond formation does not alter virus growth in vitro or in mice or ferrets (36). Chimeric BM2 (19–36aaA/M2) and BM2 d2–5 (19–36aaA/M2) ion channels were expressed in Xenopus oocytes, their channel activity and amantadine inhibition were measured by 2-electrode voltage clamp, and their properties were compared with those of WT A/M2, WT BM2, and BM2 (24–36aaA/M2) chimeric ion channels. Representative recordings (Fig. 2) and quantification of relative specific activities and percentage inhibition (Table 1) show that the BM2 (19–36aaA/M2) and the BM2 d2–5 (19–36aaA/M2) chimeric ion channels (i) had greater ion channel activity than the BM2 (24–36aaA/M2) chimeric ion channel lacking A/M2 ectodomain residues, and (ii) were inhibited by amantadine to a greater extent. However, the onset of inhibition by amantadine for the BM2 (19–36aaA/M2) and BM2 d2–5 (19–36aaA/M2) chimeric ion channels was still slower than for WT A/M2. This is most likely due to the removal of the majority of the ectodomain of A/M2, the removal of which has been shown to slow the onset of amantadine inhibition (37). This interpretation of the data is supported by the observation that the onset of inhibition of the truncated A/M2 ion channel, A/M2 (21–97), was similar to that observed for the chimeras BM2 (19–36aaA/M2) and BM2 d2–5 (19–36aaA/M2) that contain only a portion of the A/M2 ectodomain. The amantadine derivative rimantadine (100 μM) inhibited the BM2 (19–36aaA/M2) and BM2 d2–5 (19–36aaA/M2) ion channels by 85.8% ± 1.6% and 79.9% ± 0.8%, respectively, after a 5-min application.
Fig. 1.
Schematic illustration of A/M2, BM2, truncated A/M2, and chimeric BM2 proteins. The box indicates the TM domain. The A/M2 sequence is in red, and BM2 sequence is in black. Important residues are labeled with numbers corresponding to their positions in either A/M2 (red numbers) or BM2 (black numbers) proteins.
Fig. 2.
pH activation and amantadine sensitivity of A/M2, BM2, truncated A/M2, and chimeric BM2 ion channels. Representative recordings of these ion channels are shown. Amantadine sensitivity was evaluated by bathing oocytes in pH 5.5 bathing solution containing 100 μM amantadine, starting when the oocytes displayed maximum inward current (red).
Table 1.
Summary of channel properties and amantadine sensitivity
| Variable | Relative specific activity (%) | Reversal voltage (mV)* | Percentage of inhibition (%)† | IC50 (μM) amantadine‡ |
|---|---|---|---|---|
| A/M2 WT | NA | 75.5 ± 4.8 | 97.3 ± 1.0 | 16.0 ± 1.2 |
| A/M2 S31N | NA | 76.7 ± 1.5 | 31.6 ± 1.7 | 199.9 ± 13.5 |
| A/M2 (21–97) | NA | 73.2 ± 2.4 | 92.3 ± 0.7 | 45.7 ± 2.1 |
| A/M2 (21–97) S31N | NA | 74.7 ± 3.4 | 9.7 ± 3.2 | 1,554.9 ± 111.7 |
| BM2 (24–36aaA/M2) | 25.2 ± 1.8 | 75.3 ± 6.0 | 44.7 ± 2.6 | NA |
| BM2 (24–36aaA/M2) S31N | 44.7 ± 3.0 | 75.0 ± 1.5 | 5.1 ± 3.1 | NA |
| BM2 (19–36aaA/M2) | 106.0 ± 8.1 | 75.3 ± 4.7 | 83.6 ± 1.6 | 58.1 ± 3.7 |
| BM2 (19–36aaA/M2) S31N | 111.6 ± 9.1 | 76.5 ± 2.5 | 34.0 ± 1.2 | 611.1 ± 14.1 |
| BM2 d2–5 (19–36aaA/M2) | 116.4 ± 18.6 | 85.7 ± 2.2 | 73.9 ± 1.4 | 59.6 ± 4.0 |
| BM2 d2–5 (19–36aaA/M2) S31N | 109.8 ± 10.7 | 91.2 ± 1.7 | 14.0 ± 3.0 | 1,459.5 ± 86.1 |
| BM2 WT | 100.0 | 78.4 ± 0.6 | 3.7 ± 2.3 | NA |
| BM2 (37–45aaA/M2) | 73.1 ± 2.4 | 73.0 ± 2.3 | 2.6 ± 1.1 | NA |
Data are shown as mean ± SE. NA, not available
*For the measurement of reversal voltage, membrane voltage ramp measurements were made at pH 5.5 at the time when the amplitude of the steady inward current was maximal (approximately 10 s after lowering the bath solution to pH 5.5). Current–voltage relationship was plotted after subtraction of pH 8.5 background current from pH 5.5 current.
†Measurements were done after 5-min incubation with 100 μM amantadine at pH 5.5.
‡Measurements were done after 2-min incubation with amantadine at pH 5.5.
The reversal voltage of these chimeric M2 ion channels was determined to make a comparison of the ion selectivity of these chimeric ion channels with that of the WT A/M2 and WT BM2 ion channels (Table 1). The reversal voltages were very similar, indicating that these chimeric M2 ion channels are as highly proton selective as are their parent A/M2 and BM2 ion channels. Thus, an important and sensitive property of the native ion channels, ion selectivity, is preserved in the chimeric ion channels.
Introduction of the S31N Mutation into Chimeric M2 Ion Channels Reduces Amantadine Sensitivity.
As a further test of the integrity of the key properties of the chimeric ion channels, the S31N mutation was introduced into the 3 chimeric M2 proteins and A/M2 (21–97), and their amantadine sensitivity was evaluated (Fig. 2). All of the chimeric M2 proteins with the S31N mutation were very insensitive to amantadine (Table 1). Thus, the introduction of the S31N mutation into the chimeric ion channels alters amantadine sensitivity in the same way that this mutation alters the amantadine sensitivity of the WT A/M2 ion channel (Table 1).
The Chimeric BM2 (37–45aaA/M2) Ion Channel Displayed BM2-like Ion Channel Properties.
Solution NMR has indicated the presence of 4 equivalent rimantadine (amantadine) binding sites on the lipid interface of the A/M2 TM helices, with residues 41–45 interacting with rimantadine (24). To test the functional significance of this lipid-facing binding site, we transferred residues 41–45 from A/M2, together with the functionally critical residues of the HXXXW motif of A/M2, to the BM2 ion channel. The resulting chimeric BM2 ion channel, BM2 (37–45aaA/M2), was analyzed for amantadine (and rimantadine) sensitivity and other key channel properties. The chimeric BM2 ion channel, BM2 (37–45aaA/M2), was activated by pH 5.5 solution in the same way as WT BM2 and WT A/M2 channels. However, there was almost no observable effect of 100 μM amantadine on the BM2 (37–45aaA/M2) chimeric ion channel activity (Fig. 2). Because Schnell and Chou (24) had used rimantadine, this drug was also tested, and in the presence of 100 μM rimantadine BM2 (37–45aaAM2) was inhibited only 4.5% ± 1.9% (n = 3, mean ± SE). Although BM2 (37–45aaA/M2) had a slightly lower relative specific activity than WT BM2, it possessed a similar reversal voltage and therefore has ion selectivity similar to that of the WT BM2 channel (Table 1). Thus, introduction of the residues constituting the lipid-facing binding site for amantadine in the A/M2 ion channel does not impart amantadine sensitivity to the BM2 ion channel but does not significantly alter the remaining key ion channel properties of the resulting chimeric channel.
The Chimeric M2 Ion Channels Have Gating Properties That Are Indistinguishable from Those of the Parent A/M2 and BM2 Ion Channels.
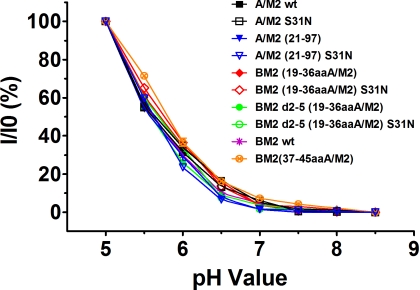
To test whether large-scale conformational changes might have occurred in the functional core of the chimeric ion channels to render them so different from their parent M2 ion channels that comparisons would be meaningless, another sensitive ion channel property, activation, was measured. To do so, the relationship between macroscopic membrane current and pHout of the chimeric M2 ion channels was compared with that of their parent A/M2 and BM2 WT ion channels. As shown in Fig. 3, the relationship between membrane current and pHout was indistinguishable among all of the constructs tested, indicating that the chimeric M2 ion channels are gated in a manner that is indistinguishable from that of their parent A/M2 WT and BM2 WT channels.
Fig. 3.
Relationship between macroscopic membrane current and pHout for oocytes expressing A/M2, BM2, truncated A/M2, and chimeric BM2 ion channel proteins. Membrane current was normalized to the value obtained at pH 5.0 (I0). Measurements were made 30 s after changing pHout from pH 8.5 to the value on the x axis.
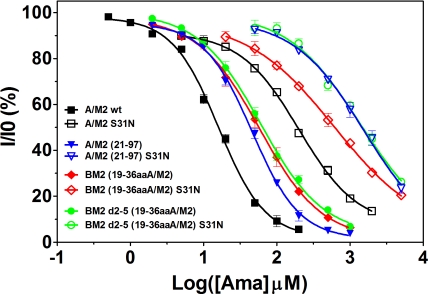
Amantadine Dose–Response Relationship of the Chimeric M2 Ion Channels and Their Corresponding S31N Mutants.
The predominant naturally occurring mutation that confers amantadine resistance to the A/M2 ion channel is A/M2 S31N. To examine the amantadine sensitivity of the chimeric channels, we modified the set of BM2 chimeric channels to contain the S31N mutation. The dose–response relationship to amantadine for the chimeric channels and their S31N derivatives was compared (Fig. 4). The IC50 values were compared with those of the WT A/M2 ion channel and the truncated A/M2 (21–97) ion channel (Table 1). For these experiments isochronic (2 min) inhibition was measured, because amantadine inhibition is only very slowly reversible for the WT A/M2 and BM2 chimeras not bearing the S31N mutation (31). The same isochronic inhibition was measured for the corresponding S31N mutants to allow a direct comparison, although the inhibition on the S31N mutants was rapidly reversible. All of the chimeric channels lacking the S31N mutation showed a saturable dose–response curve (Fig. 4). These S31N mutant channels were only inhibited by very large concentrations of amantadine, and consequently their IC50 values were much higher than those of channels without the S31N mutation. The dose–response curves were right-shifted 1–1.5 log units compared with the corresponding M2 channels without the S31N mutation. The chimeric ion channels BM2 (19–36aaA/M2) and BM2 d2–5 (19–36aaA/M2) had dose–response curves and IC50 values that were similar to that of the truncated A/M2 (21–97) ion channel. However, the dose–response curves of all 3 of these channels were slightly right-shifted when compared with the curve for the WT A/M2 ion channel. These results are consistent with the observation that the removal of the majority of the ectodomain slows the onset of amantadine inhibition by 2- to 3-fold (37). More importantly, these results demonstrate that transferring residues 19–36 from the A/M2 protein to the BM2 protein creates a chimeric ion channel with amantadine sensitivity indistinguishable from that of a truncated A/M2 (21–97) ion channel. This finding further supports the notion that the structure of the functional core of the chimeric ion channels differs from that of the parent BM2 channel in such a fashion that, among the key ion channel properties, only amantadine sensitivity differs from that of the parent BM2 ion channel. Thus, comparisons among the chimeric ion channels and their parental WT ion channels are justified, and the functionally important amantadine binding site is located within residues 19–36, a region that is readily transferable to the BM2 ion channel.
Fig. 4.
Isochronic inhibition curves for A/M2, BM2, truncated A/M2, and chimeric BM2 ion channels by amantadine. Seven to nine different concentrations of amantadine were applied to oocytes expressing the ion channels for 2 min at pH 5.5, and the current was measured before and 2 min after drug addition. Three oocytes were used for each drug concentration, and results were averaged (mean ± SE). A dose–response relationship was applied to the results using Origin 6 software, and the best-fit IC50 values for these ion channels are listed in Table 1.
Discussion
Pielak et al. (33) have questioned the validity of using oocytes as a system to test A/M2 ion channel activity, particularly if the critical D44A mutation altered ionic selectivity. We have now measured in oocytes the A/M2 D44A mutant reversal voltage (Vrev = 71.8 ± 4.0 mV, n = 5) and found it to be indistinguishable from that of WT A/M2 measured under the identical conditions, indicating that the 2 genotypes have the same proton selectivity (p = 0.57 in Student's t test). In addition, the specific activity of A/M2 D44A is 2.44 ± 0.34-fold higher than that of WT A/M2. Our view is that oocytes have proven to be a faithful heterologous expression system for ion channel recordings for many ion channels (reviewed in refs. 38 and 39). Although some small integral membrane proteins can activate endogenous chloride channels in oocytes (40), to our knowledge there is no known report of an amantadine-sensitive proton-selective channel, like A/M2 D44A, in oocytes. Furthermore, when A/M2 D44A was expressed in mammalian CHO-K1 cells, an amantadine-sensitive ion channel was recorded with properties very similar to that recorded from oocytes (34), a finding that argues against an oocyte artifact. Finally, an influenza virus reverse-engineered to contain the M2 D44A mutation is fully functional and amantadine sensitive (34). Chou and coworkers (41, 42) have performed in silico molecular dynamic simulations of the A/M2 TM domain. On the basis of their studies, these authors describe the A/M2 D44A mutant data of Jing et al. (34) thus: “the reliability of their experiments and interpretation are problematic” (41). However, this viewpoint ignores the highly reproducible biologic data, and thus the theoretical models do not conform to the observed biologic findings.
To explore the nature of the A/M2 residues that interact with amantadine, we generated a chimeric channel between amantadine-sensitive influenza A virus A/M2 and amantadine-insensitive influenza B virus BM2. We reported previously that when the residues of the outer half of the BM2 TM domain (residues 6–18) were replaced with the corresponding residues from the A/M2 TM domain (residues 24–36) the resulting chimeric BM2 ion channel [BM2 (24–36aaA/M2)] was partially sensitive (≈50%) to amantadine, with a slower onset of inhibition than for WT A/M2 (34). The data obtained with the chimeric molecules (Fig. 1) designed to improve amantadine sensitivity and define the drug-binding site experimentally yielded several important results. (i) The chimeric channels all exhibited very similar reversal voltages, indicating that they were proton selective. (ii) The relationship between the macroscopic current and pHout was indistinguishable among the constructs tested, indicating that the chimeric M2 ion channels are gated in the same way as their parental WT A/M2 and BM2 channels. These 2 findings suggest that the structurally critical regions required for normal function of the channel had not been perturbed. (iii) Amantadine inhibition of the chimeras BM2 (19–36aaA/M2) and BM2 d2–5 (19–36aaA/M2) was 84% and 74%, respectively, as compared with 92% inhibition for A/M2 (21–97), the latter being the most comparable A/M2-like construct. Rimantadine inhibited the chimeras BM2 (19–36aaA/M2) and BM2 d2–5 (19–36aaA/M2) 86% and 80%, respectively.
These data provide strong support that the amantadine/rimantadine binding site that lies within the A/M2 TM domain (residues 24–36) was transferred to the BM2 TM domain. Thus, because the drug–protein interaction is sequence specific, the data indicate that the drug-sensitive chimeric channels contain the amino acid sequence that binds the drug and that this sequence is derived from A/M2 residues 19–36. These chimeric molecules lack A/M2 residues 37–45, and thus interactions of the drug with A/M2 residues 40–45 cannot occur. Additionally, transfer of residues 37–45 did not result in amantadine sensitivity. Thus, an allosteric mechanism of drug inhibition via long-range interactions contradicts the data. It seems plausible that as amantadine partitions into lipid bilayers (43, 44), giving the drug access to the cytoplasmic side of the 4-helix bundle, the external drug binding site is an example of a second drug binding site. However, our functional data suggest that the amantadine/rimantadine binding site identified by Schnell and Chou (24) is not the primary site associated with the pharmacologic inhibition of the A/M2 ion channel.
Materials and Methods
Pasmids, mRNA Synthesis, and Microinjection of Oocytes.
All cDNA constructs were based on the genes of influenza A/Udorn/72 (A/M2) and influenza B/Lee/40 (BM2) and were cloned in pGEM-HJ. The BM2 protein and its derivatives were constructed to contain a C-terminal FLAG-tag epitope. The nucleotide sequences of all plasmid inserts were verified. The synthesis of mRNA and microinjection of oocytes have been described previously (37). The nucleotide sequence of the BM2 chimeric mRNAs was further confirmed by using AMV Super Reverse Transcriptase (Fermentas) and amplifying the product with AmpliTaq DNA Polymerase (Applied Biosystems). The nucleotide sequences, primer sequences, and PCR conditions are available upon request.
Two-Electrode Voltage Clamp Analysis.
Whole-cell 2-electrode voltage clamp currents and reversal voltage were measured 48–72 h after injection of oocytes, as described previously (37). Briefly, individual oocytes were held at a voltage of −20 mV and bathed in normal Barth's solution [88.0 mM NaCl, 1.0 mM KCl, 2.4 mM NaHCO3, 0.3 mM NaNO3, 0.71 mM CaCl2, 0.82 mM MgSO4, and 15 mM Hepes (for pH 8.5) or 15 mM Mes (for pH 5.5)]. For the measurement of reversal voltage, membrane voltage ramp measurements were made at pH 5.5 at the time when the amplitude of the steady inward current was maximal (approximately 10 s after lowering the bath solution to pH 5.5). The membrane voltage ramp measurements were also made at pH 8.5. The voltage ramp spanned a 110-mV range, from −50 to 60 mV. Current–voltage relationship was plotted after subtraction of pH 8.5 background current from pH 5.5 current. Currents were acquired and analyzed using the pCLAMP 10.0 software package (Axon Instruments).
Immunofluorescence of Living Oocytes and the Calculation of Relative Specific Activity.
The detailed procedure of immunofluorescence of living oocytes was described previously (13). The relative specific activity of the FLAG-tagged BM2 channel and its derivative chimeric channels was calculated from the ratio of steady current that flowed at pH 5.5 to the relative amount of protein that was expressed at the surface of the oocyte for each cell studied. For each experiment, at least 5 oocytes expressing the BM2 protein and 3 uninjected oocytes were also measured. The whole-cell membrane current for a series of individual oocytes was plotted as a function of surface expression measured in the same single oocyte. The relative specific activity of a given chimeric BM2 ion channel was calculated by normalizing the slope for chimeric BM2 ion channels to the slope of WT BM2 protein plot.
Acknowledgments.
This research was supported by National Institutes of Health Grants R01 AI-20201 (to R.A.L.) and R01 Al-57363 (to L.H.P.). R.A.L. is an Investigator of the Howard Hughes Medical Institute.
Footnotes
The authors declare no conflict of interest.
References
- 1.Lamb RA, Zebedee SL, Richardson CD. Influenza virus M2 protein is an integral membrane protein expressed on the infected-cell surface. Cell. 1985;40:627–633. doi: 10.1016/0092-8674(85)90211-9. [DOI] [PubMed] [Google Scholar]
- 2.Paterson RG, Takeda M, Ohigashi Y, Pinto LH, Lamb RA. Influenza B virus BM2 protein is an oligomeric integral membrane protein expressed at the cell surface. Virology. 2003;306:7–17. doi: 10.1016/s0042-6822(02)00083-1. [DOI] [PubMed] [Google Scholar]
- 3.Pinto LH, Holsinger LJ, Lamb RA. Influenza virus M2 protein has ion channel activity. Cell. 1992;69:517–528. doi: 10.1016/0092-8674(92)90452-i. [DOI] [PubMed] [Google Scholar]
- 4.Chizhmakov IV, et al. Selective proton permeability and pH regulation of the influenza virus M2 channel expressed in mouse erythroleukaemia cells. J Physiol. 1996;494:329–336. doi: 10.1113/jphysiol.1996.sp021495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mould JA, et al. Influenza B virus BM2 protein has ion channel activity that conducts protons across membranes. Dev Cell. 2003;5:175–184. doi: 10.1016/s1534-5807(03)00190-4. [DOI] [PubMed] [Google Scholar]
- 6.Holsinger LJ, Lamb RA. Influenza virus M2 integral membrane protein is a homotetramer stabilized by formation of disulfide bonds. Virology. 1991;183:32–43. doi: 10.1016/0042-6822(91)90115-r. [DOI] [PubMed] [Google Scholar]
- 7.Sugrue RJ, Hay AJ. Structural characteristics of the M2 protein of the influenza A viruses: Evidence that it forms a tetrameric channel. Virology. 1991;180:617–624. doi: 10.1016/0042-6822(91)90075-M. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sakaguchi T, Tu Q, Pinto LH, Lamb RA. The active oligomeric state of the minimalistic influenza virus M2 ion channel is a tetramer. Proc Natl Acad Sci USA. 1997;94:5000–5004. doi: 10.1073/pnas.94.10.5000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pinto LH, et al. A functionally defined model for the M2 proton channel of influenza A virus suggests a mechanism for its ion selectivity. Proc Natl Acad Sci USA. 1997;94:11301–11306. doi: 10.1073/pnas.94.21.11301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bauer CM, Pinto LH, Cross TA, Lamb RA. The influenza virus M2 ion channel protein: Probing the structure of the transmembrane domain in intact cells by using engineered disulfide cross-linking. Virology. 1999;254:196–209. doi: 10.1006/viro.1998.9552. [DOI] [PubMed] [Google Scholar]
- 11.Shuck K, Lamb RA, Pinto LH. Analysis of the pore structure of the influenza A virus M(2) ion channel by the substituted-cysteine accessibility method. J Virol. 2000;74:7755–7761. doi: 10.1128/jvi.74.17.7755-7761.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Balannik V, Lamb RA, Pinto LH. The oligomeric state of the active BM2 ion channel protein of influenza B virus. J Biol Chem. 2008;283:4895–4904. doi: 10.1074/jbc.M709433200. [DOI] [PubMed] [Google Scholar]
- 13.Ma C, et al. Identification of the pore-lining residues of the BM2 ion channel protein of influenza B virus. J Biol Chem. 2008;283:15921–15931. doi: 10.1074/jbc.M710302200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mould JA, et al. Mechanism for proton conduction of the M2 ion channel of influenza A virus. J Biol Chem. 2000;275:8592–8599. doi: 10.1074/jbc.275.12.8592. [DOI] [PubMed] [Google Scholar]
- 15.Hu J, et al. Histidines, heart of the hydrogen ion channel from influenza A virus: Toward an understanding of conductance and proton selectivity. Proc Natl Acad Sci USA. 2006;103:6865–6870. doi: 10.1073/pnas.0601944103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Venkataraman P, Lamb RA, Pinto LH. Chemical rescue of histidine selectivity filter mutants of the M2 ion channel of influenza A virus. J Biol Chem. 2005;280:21463–21472. doi: 10.1074/jbc.M412406200. [DOI] [PubMed] [Google Scholar]
- 17.Tang Y, Zaitseva F, Lamb RA, Pinto LH. The gate of the influenza virus M2 proton channel is formed by a single tryptophan residue. J Biol Chem. 2002;277:39880–39886. doi: 10.1074/jbc.M206582200. [DOI] [PubMed] [Google Scholar]
- 18.Sugrue RJ, et al. Specific structural alteration of the influenza haemagglutinin by amantadine. EMBO J. 1990;9:3469–3476. doi: 10.1002/j.1460-2075.1990.tb07555.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hay AJ. The action of adamantanamines against influenza A viruses: Inhibition of the M2 ion channel protein. Semin Virol. 1992;3:21–30. [Google Scholar]
- 20.Lamb RA, Holsinger LJ, Pinto LH. The influenza A virus M2 ion channel protein and its role in the influenza virus life cycle. In: Wimmer E, editor. Receptor-Mediated Virus Entry into Cells. Cold Spring Harbor, NY: Cold Spring Harbor Lab Press; 1994. pp. 303–321. [Google Scholar]
- 21.Davies WL, et al. Antiviral activity of 1-adamantanamine (amantadine) Science. 1964;144:862–863. doi: 10.1126/science.144.3620.862. [DOI] [PubMed] [Google Scholar]
- 22.Hay AJ, Wolstenholme AJ, Skehel JJ, Smith MH. The molecular basis of the specific anti-influenza action of amantadine. EMBO J. 1985;4:3021–3024. doi: 10.1002/j.1460-2075.1985.tb04038.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stouffer AL, et al. Structural basis for the function and inhibition of an influenza virus proton channel. Nature. 2008;451:596–599. doi: 10.1038/nature06528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schnell JR, Chou JJ. Structure and mechanism of the M2 proton channel of influenza A virus. Nature. 2008;451:591–595. doi: 10.1038/nature06531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kovacs FA, Cross TA. Transmembrane four-helix bundle of influenza A M2 protein channel: Structural implications from helix tilt and orientation. Biophys J. 1997;73:2511–2517. doi: 10.1016/S0006-3495(97)78279-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tian C, Tobler K, Lamb RA, Pinto LH, Cross TA. Expression and initial structural insights from solid-state NMR of the M2 proton channel from influenza A virus. Biochemistry. 2002;41:11294–11300. doi: 10.1021/bi025695q. [DOI] [PubMed] [Google Scholar]
- 27.Li C, Qu H, Gao FP, Cross TA. Solid-state NMR characterization of conformational plasticity within the transmembrane domain of the influenza A M2 proton channel. Biochim Biophys Acta. 2007;1768:3162–3170. doi: 10.1016/j.bbamem.2007.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hu J, et al. Backbone structure of the amantadine-blocked trans-membrane domain M2 proton channel from Influenza A virus. Biophys J. 2007;92:4335–4343. doi: 10.1529/biophysj.106.090183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cady SD, Mishanina TV, Hong M. Structure of amantadine-bound M2 transmembrane peptide of influenza A in lipid bilayers from magic-angle-spinning solid-state NMR: The role of ser31 in amantadine binding. J Mol Biol. 2009;385:1127–1141. doi: 10.1016/j.jmb.2008.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lamb RA, Pinto LH. The proton selective ion channels of influenza A and B viruses. In: Kawaoka Y, editor. Contemporary Topics in Influenza Virology. Wymondham, Norfolk, UK: Horizon Scientific Press; 2005. pp. 65–93. [Google Scholar]
- 31.Wang C, Takeuchi K, Pinto LH, Lamb RA. Ion channel activity of influenza A virus M2 protein: Characterization of the amantadine block. J Virol. 1993;67:5585–5594. doi: 10.1128/jvi.67.9.5585-5594.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tang Y, Venkataraman P, Knopman J, Lamb RA, Pinto LH. The M2 proteins of influenza A and B viruses are single-pass proton channels. In: Fischer WB, editor. Viral Membrane Proteins: Structure, Function and Drug Design. Amsterdam: Kluwer Academic; 2005. pp. 101–111. [Google Scholar]
- 33.Pielak RM, Schnell JR, Chou JJ. Mechanism of drug inhibition and drug resistance of influenza A M2 channel. Proc Natl Acad Sci USA. 2009;106:7379–7384. doi: 10.1073/pnas.0902548106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jing X, et al. Functional studies indicate amantadine binds to the pore of the influenza A virus M2 proton-selective ion channel. Proc Natl Acad Sci USA. 2008;105:10967–10972. doi: 10.1073/pnas.0804958105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lin TI, Schroeder C. Definitive assignment of proton selectivity and attoampere unitary current to the M2 ion channel protein of influenza A virus. J Virol. 2001;75:3647–3656. doi: 10.1128/JVI.75.8.3647-3656.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Castrucci MR, et al. The cysteine residues of the M2 protein are not required for influenza A virus replication. Virology. 1997;238:128–134. doi: 10.1006/viro.1997.8809. [DOI] [PubMed] [Google Scholar]
- 37.Ma C, et al. Identification of the functional core of the influenza A virus A/M2 proton-selective ion channel. Proc Natl Acad Sci USA. 2009;106:12283–12288. doi: 10.1073/pnas.0905726106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Smart TG, Krishek BJ. Xenopus oocyte microinjection and ion channel expression. In: Boulton A, Baker G, Walz W, editors. Neuromethods. Patch-Clamp Applications and Protocols. Vol 26. New York: Humana; 1995. pp. 259–305. [Google Scholar]
- 39.Goldin AL. Expression of ion channels in Xenopus oocytes. In: Clare JJ, Trezise DJ, editors. Expression and Analysis of Recombinant Ion Channels. Weinheim, Germany: Wiley-VCH; 2006. pp. 1–25. [Google Scholar]
- 40.Shimbo K, Brassard DL, Lamb RA, Pinto LH. Viral and cellular small integral membrane proteins can modify ion channels endogenous to Xenopus oocytes. Biophys J. 1995;69:1819–1829. doi: 10.1016/S0006-3495(95)80052-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Du QS, Huang RB, Wang CH, Li XM, Chou KC. Energetic analysis of the two controversial drug binding sites of the M2 proton channel in influenza A virus. J Theoretical Biol. 2009;259:159–164. doi: 10.1016/j.jtbi.2009.03.003. [DOI] [PubMed] [Google Scholar]
- 42.Huang RB, Du QS, Wang CH, Chou KC. An in-depth analysis of the biological functional studies based on the NMR M2 channel structure of influenza A virus. Biochem Biophys Res Commun. 2008;377:1243–1247. doi: 10.1016/j.bbrc.2008.10.148. [DOI] [PubMed] [Google Scholar]
- 43.Wang J, Schnell JR, Chou JJ. Amantadine partition and localization in phospholipid membrane: A solution NMR study. Biochem Biophys Res Commun. 2004;324:212–217. doi: 10.1016/j.bbrc.2004.09.039. [DOI] [PubMed] [Google Scholar]
- 44.Epand RM, Epand RF, McKenzie RC. Effects of viral chemotherapeutic agents on membrane properties: Studies of cyclosporin A, benzlooxycarbonyl-D-Phe-L-Phe-Gly and amantadine. J Biol Chem. 1987;262:1526–1529. [PubMed] [Google Scholar]