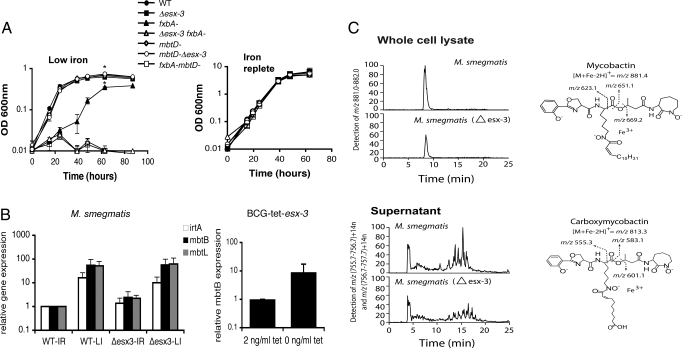
Fig. 3.
Esx-3 interacts with the mycobactin pathway but is not required for sensing iron or for siderophore synthesis. (A) Growth of wild-type (WT), Δesx-3, exochelin (fxbA−), and mycobactin (mbtD−) insertional biosynthesis mutants and double mutants (fxbA− Δesx-3 and fxbA− mbtD−) in low iron or iron-replete Sauton's medium (n = 3; *, P < 0.001). Strains were not subcultured before inoculation. (B) Quantitative RT-PCR for mbtB, mbtL, and irtA expression in WT and Δesx-3 Mycobacterium smegmatis (n = 3) in low iron (LI) or iron-replete (IR) medium and for mbtB in bacillus Calmette–Guérin (BCG)-tet-esx-3 in low iron medium. Data representative of three independent experiments are shown. Error bars represent the standard deviations. (C) The LC-MS analysis of lipids extracted from the whole cell lysates of M. smegmatis with chlorofrom and methanol yielded mycobactin chromatograms in which the m/z (881.4), elution time, and fragments (right) correspond to the expected and actual measurements of an authentic standard. Conditioned supernatants were subjected to LC-MS detection in mass windows corresponding to the known masses of carboxymycobactins [m/z (756.7–757.7) + 14n, where n is 0, 1, 2, 3, 4] and [m/z (757.7–758.7) + 14n] (upper). The elution time and collision-induced dissociation MS fragmentation (right) of the putative carboxymycobactins are consistent with those of an authentic carboxymycobactin standard.