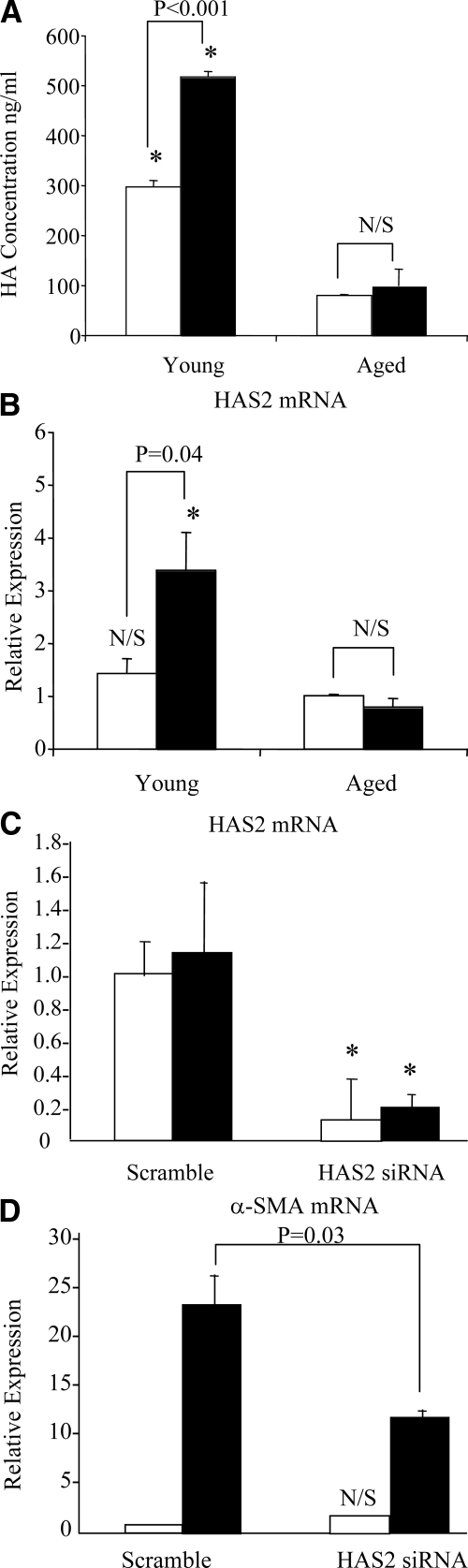
Figure 4.
Effect of age on HAS2 mRNA expression and its involvement in TGF-β1-dependent fibroblast activation. A and B: Confluent monolayers of patient-matched young and aged dermal fibroblasts were growth-arrested in serum-free medium for 48 hours. The medium was then replaced with either serum-free medium alone (white bars) or serum-free medium containing 10 ng/ml TGF-β1 (black bars), and the incubations were continued for 72 hours. HA secreted into the culture medium was quantified by ELISA. After removal of the culture medium, total mRNA was extracted and cDNA prepared as described in Materials and Methods. HAS2 expression was assessed by RT-QPCR. Ribosomal RNA expression was used as an endogenous control, and gene expression was assessed relative to control aged fibroblasts. The comparative CT method was used for relative quantification of gene expression. Data are presented as the mean ± SE of six individual experiments using cells from two patient donors. Statistical analysis was performed by Student’s t-test: #not significant; *P < 0.01, compared with aged cells. N/S, not significant. C and D: To further examine the role of HAS2, young dermal fibroblasts were transfected with HAS2 siRNA or scrambled oligonucleotide control (scramble) and incubated in medium supplemented with 10% FBS for 24 hours. The medium was then replaced with serum-free medium for a further 24 hours, before addition of serum-free medium alone (white bars) or serum-free medium containing 10 ng/ml TGF-β1 (black bars) for 72 hours. Total mRNA was extracted and HAS2 (C) and α-SMA (D) expression was assessed by RT-QPCR. Ribosomal RNA expression was used as an endogenous control, and gene expression was assessed relative to control scramble samples. The comparative CT method was used for relative quantification of gene expression, and the results represent the mean ± SE of nine individual experiments using cells isolated from three different donors. Statistical analysis was performed by the Student’s t-test: N/S, not significant; *P < 0.01, compared with scramble.