Abstract
We have compared the physical properties of a 15.51 Kbp constitutive heterochromatin segment and a 16.17 Kbp facultative heterochromatin segment that form part of the chicken β-globin locus. These segments were excised from an avian erythroleukemia cell line by restriction enzyme digestion and released from the nucleus, thus allowing measurement of the sedimentation coefficients using calibrated sucrose gradients. A determination of the buoyant density of the cross-linked particle in CsCl led to the total mass of the particles, and their frictional coefficients, f. Despite the slight differences in nucleosome density, the measured value of f for both fragments was consistent with a rod like particle having a diameter of 33 – 45 nm and a length corresponding to approximately 6 – 7 nucleosomes per 11 nm turn. At higher ionic strengths we found no evidence of any abrupt conformational change, demonstrating that these chromatin fragments released from the nucleus did not assume the more compact conformations recently described for some reconstituted structures.
Keywords: chromatin, structure, hydrodynamics, heterochromatin
Introduction
Although the structure of the nucleosome is well understood, less is known about the folding of polynucleosome chains to form higher order chromatin structures within the nucleus. Most of the physical studies on particles obtained by partial nuclease digestion indicate that the first order of folding involves the packing of nucleosomes into helical arrays to form relatively rigid, rod like structures with a diameter of about 30 nm (the ‘30 nm fiber’). The evidence for these properties comes from electron microscopy,1,2 X-ray3,4,5,6 and neutron scattering,7,8,9 as well as hydrodynamic measurements.10,11,12,13 Careful correlation of neutron scattering and electron microscopic results shows that chromatin released from nuclei has a mass per unit length that increases with salt concentration, but reaches a plateau value, corresponding to slightly more than 6 nucleosomes per 11 nm of length, in the range of 75 – 125 mM NaCl.8 This of course reflects the average properties of the chromatin released from the nucleus, and is likely to be dominated by contributions from condensed chromatin containing closely packed nucleosomes.
Most of this chromatin contains non-coding DNA sequences, including repetitive DNAs that have no long open reading frames. Recent results suggest that such regions may be transcribed at very low levels, perhaps part of a mechanism that maintains them in their condensed state. The condensed chromatin fraction might also contain regions coding for genes that are not transcriptionally active in the cells from which they are isolated. There has been relatively little information about the chromatin conformation assumed by such silent genes14,15,16 and repetitive DNA sequences,17 but a recent and rather extensive examination shows that the sedimentation properties of gene-rich euchromatin and centromeric heterochromatin differ, such that the latter adopts a relatively compact conformation.18
Measurements of sedimentation properties alone provide only ambiguous information about the shape of a particle, since the sedimentation coefficient depends upon both mass and shape. For that reason we undertook, in an earlier paper, a study of a single, well-defined fragment containing 15.51 Kbp of DNA derived from the constitutively condensed chromatin fragment immediately upstream of the chicken β-globin locus.19 We combined precise determination of the particle’s sedimentation coefficient with measurements of its buoyant density, allowing us to calculate its frictional coefficient, and to show that this was consistent with the expected properties of a rod like structure with a diameter of 33 – 45 nm and a mass per unit length of approximately 5 – 6 nucleosomes per 11 nm, dimensions similar to those described above for the 30 nm fiber.
This can be compared with results of physical and structural studies in the Richmond laboratory of polynucleosomes reconstituted from bacterially expressed histones and DNA of defined sequence. X-ray diffraction measurements of a tetranucleosome array at 9 A˚́ resolution show a two-start helical structure,20 consistent with earlier measurements on a 12 nucleosome repeat structure.21 Using this as a guide to stack multiple copies of the tetranucleosome on one another, the authors are able to construct a model fiber with a diameter of 24 – 25 nm and a repeat of 5.8 –6.6 nucleosomes per 11 nm of length. A quite different set of rod like structures emerges, however, from electron microscopic studies of reconstitutes containing between 47 and 72 nucleosomes bound to DNA composed of sequence repeats with the same strong histone octamer positioning properties.22 The structural details vary with the length of the individual repeats; for shorter repeat lengths (177 to 207 bp) the rod has a diameter of about 35 nm, but the observed mass per unit length is about 10 nucleosomes per 11 nm of length, much larger than the values discussed above. Even higher compaction is observed at longer repeat lengths. It is suggested by these authors that this degree of compaction, greater than any previously observed, may reflect the use of higher salt concentrations than those used in other studies.
The problem of reconciling these disparate results for reconstituted chromatin is separate from the question of how they compare with the properties of chromatin fragments isolated from living cells. For this reason we turned again to the chromatin of the chicken β-globin locus, which, as mentioned above, contains a well defined heterochromatic structure. It also contains, adjacent to this constitutively silent region, an extended sequence at least that long containing members of the β-globin gene family. The cells we use are at a stage of erythroid development where these genes are not yet expressed (or only at a low level), and where they are not marked by histone modifications associated with transcriptionally active chromatin.23,24 This provides the opportunity to compare the properties of the two regions within the same cell. Applying the methods we had used earlier,19 we show that this region of facultative heterochromatin is quite similar in hydrodynamic properties to the constitutive heterochromatin immediately upstream. We also extend the study of both regions to higher ionic strength solvents, and find no evidence, in these naturally formed chromatin structures, of any abrupt conformational change. In every case, the hydrodynamic data are consistent with structures within the range we had previously described, with a mass per unit length of about 6 nucleosomes per 11 nm. These chromatin fragments, at least, do not assume the more compact conformations reported for some reconstituted structures.
Results
Hydrodynamic analysis of a 16.17 Kbp β-globin gene fragment
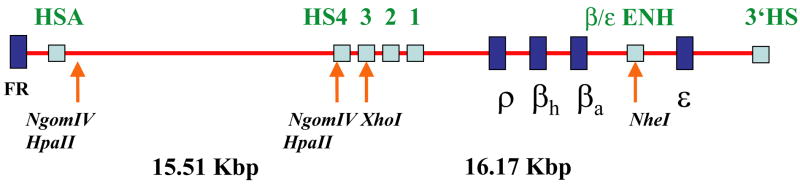
A previous study of the 15.51 Kbp condensed chromatin fragment released by HpaII digestion of nuclei prepared from 6C2 cells revealed the presence of a homogenous chromatin fragment. The sedimentation properties obtained using a calibrated sucrose gradient, combined with the molecular mass based on the particle buoyancy in a CsCl gradient, allowed for the determination of the frictional coefficient f. This value is consistent with a rod like particle of the approximate length and diameter proposed earlier for the 30 nm chromatin fiber.1 To further investigate the chromatin fiber structure, the physical properties of the 15.51 Kbp constitutive heterochromatin fragment were compared to those of a similarly sized chromatin fragment spanning the β-globin gene locus. Earlier studies of the chicken β-globin locus in 6C2 cells have shown that the βA/ε globin gene enhancer is hypersensitive and accessible to digestion with the endonuclease NheI,25 this representing a unique restriction site within the ~ 55 Kbp spanning the whole locus shown in Figure 1. An analysis of this sequence26 also identifies a series of four unique XhoI sites. Two XhoI sites are found upstream within the folate receptor gene; another site is found at 8.679 Kbp, within the condensed chromatin fragment. This condensed fragment is highly methylated at CpG sites,26 as well as resistant to digestion with restriction enzymes.19 The remaining site located at 25.743 Kbp is within hypersensitive site 3 (HS3) of the locus control region and was expected to be accessible in 6C2 nuclei. A digest with NheI and XhoI would therefore yield a 16.17 Kbp β-globin chromatin fragment. Indeed, a Southern blot analysis of DNA obtained from 6C2 nuclei digested with XhoI and NheI revealed the release of a 16.17 Kbp β-globin chromatin fragment, and confirmed the inaccessibility of the three upstream XhoI sites to digestion (data not shown). To compare directly the properties of the 16.17 Kbp β-globin chromatin fragment with those of the 15.51 Kbp condensed chromatin fragment two unique NgomIV restriction sites, representing subsets of the HpaII sites demarcating the condensed chromatin region, were used (Figure 1). Unlike HpaII, no such restriction sites are present within the rest of the locus.
Figure 1.
Diagram of the chicken β-globin locus, showing the 15.51 Kbp condensed chromatin region demarcated by HpaII or NgomIV and the 16.17 Kbp β-globin region flanked by XhoI and NheI.26 These represent the constitutive and facultative heterochromatin fragments, respectively. Hypersensitive sites spanning the ~55 Kbp sequence are indicated as light blue boxes, as is the β/ε enhancer. The dark blue boxes indicate the developmentally regulated folate receptor (FR) and β-globin genes. The restrictions sites accessible in 6C2 nuclei are highlighted.
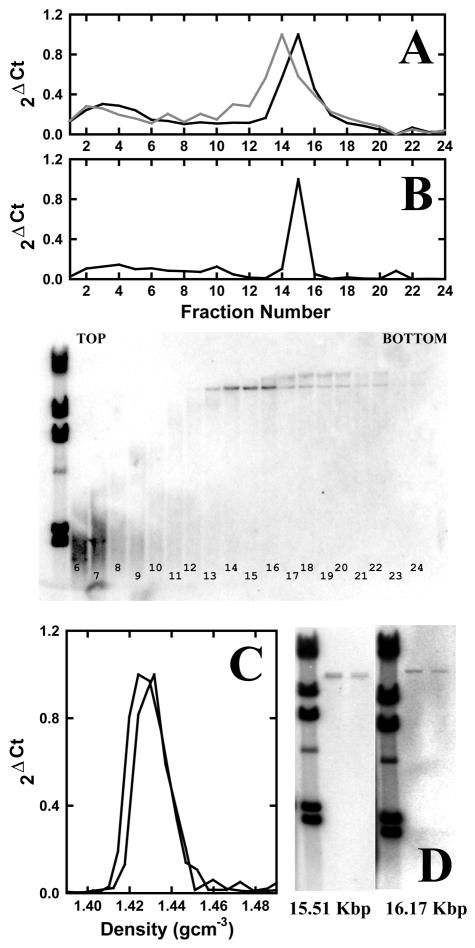
Nuclei from chicken 6C2 cells were prepared, digested with a combination of NgomIV, XhoI and NheI, and dialyzed into a buffer that released the digestion products from the nuclei. Typically, 20–30% of the input genomic DNA was released, and the specific fragments were present approximately in genomic abundance in this released fraction as determined by quantitative PCR. Following sucrose gradient sedimentation and DNA purification, the fractions collected were analyzed by quantitative PCR using probes for both the 15.51 Kbp condensed chromatin and the 16.17 Kbp β-globin fragments. The PCR method revealed a single peak for both fragments (Figure 2A), and the sedimentation properties of the 15.51 Kbp NgomIV condensed chromatin fragment were indistinguishable from those obtained for the identical HpaII fragments.19 In addition, the 16.17 Kbp XhoI/NheI β-globin fragment reproducibly sedimented as a single peak, one fraction further than the condensed chromatin fragment. Similar observations were made when 6C2 nuclei were digested only with XhoI and NheI (Figure 2B). To confirm both the size and integrity of the fragment, individual fractions obtained by sucrose gradient sedimentation were analyzed by Southern analysis using a radioactively labeled probe uniquely targeted to 16.17 Kbp XhoI/NheI β-globin fragment. As illustrated in Figure 2B, the fragment detected by PCR corresponds to the approximately 16 Kbp fragment observed in the Southern analysis. Furthermore, a PCR analysis using various probes spanning the length of the fragment led to profiles similar to those observed in Figure 2B (data not shown). Using the sucrose gradient calibration previously established,19 we concluded that the 16.17 Kbp XhoI/NheI β-globin chromatin fragment sediments with an s20,w of 138 ± 2 S.
Figure 2.
Sucrose gradient ultracentrifugation of the 15.5 Kbp NgomIV and 16.17 Kbp XhoI and NheI fragments. (A) Samples containing the NgomIV, XhoI and NheI nuclear digest were centrifuged on a 5 – 30% (w/v) sucrose gradient in DB-80 (80 mM NaCl) for 1.5 hours at 38,000 rpm (SW40-Ti rotor). Sucrose gradient fractions were collected and the DNA from each fraction was isolated and the 15.51 and 16.17 Kbp fragments were detected by quantitative PCR using probes specific for each chicken sequence. The ordinate axis, expressed in terms of a normalized 2(ΔCt), is proportional to the concentration of the DNA of interest. The PCR method shows that the 16.17 Kbp β-globin fragment (black, probe 27.64) migrates one fraction further into the gradient than the 15.51 Kbp condensed chromatin fragment (grey, probe 17.763). (B) Samples containing the XhoI and NheI nuclear digest were centrifuged on a sucrose gradient as in (A). The DNA from each fraction was isolated, resolved by agarose gel electrophoresis, and analyzed by Southern blotting using probe 410, specific for the 16.17 Kbp β-globin fragment. Fractions are numbered and the markers on the left represent a labeled λ/HindIII digest. The blot reveals that the 16.17 Kbp chromatin fragment sediments as a homogeneous component. The corresponding quantitative PCR analysis with probe 27.649 for this gradient is shown above the Southern blot. (C) Banding profiles (from two experiments) of the crosslinked 16.17 Kbp chromatin fragment in a CsCl gradient as analyzed by quantitative PCR. The gradient was formed in a TLS-55 rotor at 20.0°C and 30,000 rpm for 96 hours. (D) Pooled DNA was prepared from the CsCl fractions and analyzed by Southern blotting using probes specific for the 15.51 Kbp condensed fragment and the 16.17 Kbp β-globin fragment. The markers on the left represent a labeled λ/HindIII digest.
To determine the molecular mass of this 16.17 Kbp XhoI/NheI β-globin chromatin fragment, the peak gradient fraction was cross-linked with formaldehyde and its buoyant density was determined in a CsCl gradient.16,19 PCR was used to detect the specific particle and, unlike the 15.51 Kbp condensed chromatin, this fragment was paucidisperse spanning a density range of 1.41 to 1.45 gcm−3 (Figure 2C, 2D), as typically observed for chromatin.17,27 The buoyant density distribution had a maximum at 1.424 gcm−3; based on this and the known DNA content of the particle, a molecular mass of 22.0 MDa was calculated. This value is slightly smaller than the mass of 23.3 MDa previously determined for the 15.51 Kbp condensed chromatin fragment. If the protein component consists entirely of histones, then these results reflect a decreased histone content corresponding to an increased repeat length of 186 bp, and a 20 bp linker. Whereas the 15.51 Kbp condensed chromatin fragment is found to have 89 nucleosomes,19 the 16.17 Kbp XhoI/NheI β-globin chromatin fragment has an average of 87 nucleosomes. If proteins other than histones are also bound, then the number of nucleosomes on the β-globin fragment must be even lower. This fragment thus has as an upper limit nucleosome density about 6% lower than that of the 15.51 Kbp condensed chromatin fragment.
A frictional coefficient f of 0.96 × 10−6 gs−1 for the 16.17 Kbp XhoI/NheI β-globin chromatin fragment was calculated from the sedimentation and buoyant density data. As described for the 15.51 Kbp condensed chromatin fragment,19,28,29 this frictional coefficient corresponds to rod lengths accommodating 6 – 7 nucleosomes per turn for diameters of 33 – 45 nm. However, unlike the condensed chromatin fragment, which is a close packed polynucleosome structure based both on its buoyant density and observed resistance to MspI digestion,19 the 16.17 Kbp β-globin fragment may not be as tightly packed. This was confirmed by BamHI digestion of 6C2 nuclei which yields all of the three β-globin fragments expected, as evidenced by Southern blotting and quantitative PCR (data not shown).
The XhoI and NheI restriction sites, located within erythroid specific hypersensitive sites, provide a method to release the developmentally regulated β-globin gene chromatin fragment. In 10-day erythrocytes the β-globin locus possesses all of the hallmarks of active chromatin23,24 consistent with the known expression of the βA gene.25 Although we wanted to compare the properties of the facultative heterochromatin fragment released from 6C2 nuclei with the same fragment from 10-day chicken erythrocyte nuclei, a sequence polymorphism, quite common in chicken, has prevented us from doing so: the DNA in these nuclei contains an additional XhoI or NheI restriction site within the 16.17 Kbp β-globin fragment (data not shown).
Ionic strength dependence and comparison with hydrodynamic theory
Studies on the condensation of chromatin as a function of the ionic strength show that the polynucleosome chain adopts a loose filamentous structure having a diameter of 10 nm at low ionic strength.1 Hydrodynamic,10,11,12,13 X-ray5 and neutron scattering7,8 studies all support the condensation of chromatin into a 30 nm fiber as the ionic strength is increased. Whereas the hydrodynamic studies all show a monotonous increase in the sedimentation coefficient as the monovalent NaCl concentration is increased to 120 mM, neutron scattering studies on long chromatin (~ 22 Kbp) demonstrate that the mass per unit length reaches a constant value at 75 mM NaCl or 40 mM NaCl and 0.25 mM MgCl2.8 Unlike the sedimentation coefficients, the mass per unit length measured by small angle neutron scattering is not influenced significantly by ‘breaks’ in the fiber.
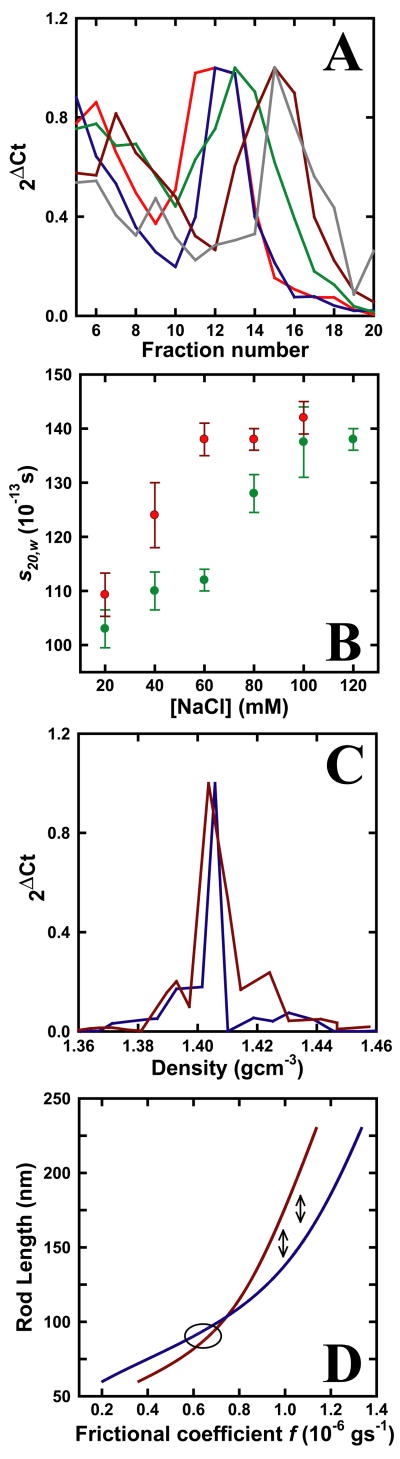
Recent electron microscopic studies carried out using reconstitutes on 601 positioning repeat sequences31 containing between 47 and 72 nucleosomes reveal a mass per unit length of about 10 nucleosomes per 11 nm, in 1.0 to 1.6 mM MgCl2.22 These values are much larger than the usual 6 – 7 nucleosomes per 11 nm and it was suggested that this greater degree of compaction may reflect the use of the higher divalent cation concentrations. Based on the sedimentation properties of similar reconstitutes on somewhat different repeat sequences containing 12 and 19 nucleosomes, Huynh and coworkers32 showed that the sedimentation coefficients obtained in 120 mM NaCl are essentially identical to those obtained in 1.0 mM MgCl2. The ionic strength dependence of the sedimentation coefficient for these reconstitutes is, for all intents and purposes, identical to that observed for similarly sized chromatin obtained from rat liver nuclei..32, and the sedimentation properties correspond to the 6–7 nucleosome per 11 nm model. We wanted to determine whether raising the salt concentration would lead in the case of our fragments, derived from nuclei, to the formation of more highly compact structures related to those observed by Robinson et al.22 To that end, the sedimentation coefficients of both the 15.51 Kbp and 16.17 Kbp heterochromatin fragments were determined as a function of ionic strength. Nuclei from chicken 6C2 cells were prepared as above, and digested with HpaII, or a combination of XhoI and NheI, to release the 15.51 Kbp condensed or 16.17 Kbp β-globin chromatin fragments, respectively. Digests were dialyzed into a buffer containing 80 mM NaCl and the soluble digestion products were layered on 5 – 30% sucrose gradients prepared in 10 mM Tris (pH = 7.4), 0.2 mM EDTA and various NaCl concentrations. Following sucrose gradient sedimentation and DNA purification, the fractions collected were analyzed by quantitative PCR as described above. In all cases the PCR method detects a single peak and for a given chromatin preparation, the fraction number corresponding to the peak increases with the ionic strength. Representative data obtained for the 15.51 Kbp HpaII condensed chromatin fragment are shown in Figure 3A. Using the previous calibration,19 the sedimentation coefficients were thus determined as a function of the ionic strength (Figure 3B). As the densities and viscosities of 20 to 120 mM NaCl solutions do not vary by more than one percent at 4°C, the sedimentation coefficients are primarily determined by the sucrose concentration and fraction number, justifying the use of the calibration made in 80 mM NaCl. Furthermore, all the measured sedimentation coefficients are interpolated. The values determined for both the chromatin fragments (Figure 3B) are within the range of those published for similarly sized fragments at various ionic strengths.12
Figure 3.
Ionic strength dependence of the sedimentation coefficients. (A) Samples containing the HpaII nuclear digest were centrifuged on sucrose gradients at different ionic strengths. DNA from each fraction was isolated and detected by quantitative PCR using the 13.192 or 17.763 probes specific for the 15.51 Kbp condensed chromatin. The PCR method shows that the 15.51 Kbp chromatin fragment migrates further into the gradient as the ionic strength is increased. Data are representative for those observed in 20 (red), 40 (blue), 60 (green), 100 (brown) and 120 (grey) mM NaCl. (B) A plot of the sedimentation coefficient as a function of the ionic strength for both the 15.51 Kbp condensed chromatin fragment (green) and the 16.17 Kbp β-globin fragment (red). The peak maxima shown in (A) were used to obtain the sedimentation coefficient based on the calibrated sucrose gradients.19 Data are the average of at least two (16.17 Kbp) or three (15.51 Kbp) experiments. (C) Banding profiles of the crosslinked 15.51 Kbp chromatin fragment in a TLS-55 CsCl gradient as analyzed by quantitative PCR. DNA was prepared from sucrose gradients carried out in 20 (blue) and 120 mM NaCl (red). (D) Predicted relationship based on the theory of Broersma28,29 between the rod length and frictional coefficient for rod like particles having diameters of either 33 (red line) or 45 nm (blue line). The calculated frictional coefficients in 80mM (right double arrow) and 120mM NaCl (left double arrow) solvents for the 15.51 Kbp heterochromatin fragment are indicated by double arrows. The range of predicted frictional coefficients for the more compact structure of Robinson et al.22 is indicated by an ellipse.
To confirm that the molecular mass of the 15.51 Kbp condensed chromatin fragment remained unchanged following sedimentation at 20 or 120 mM NaCl, sucrose gradient fractions containing the chromatin fragment were collected, formaldehyde cross-linked and analyzed on CsCl gradients (Figure 3C). Irrespective of the ionic strength, the cross-linked chromatin was found to band at a density identical to the one previously determined.19 However, the sample recovered from the 120 mM NaCl sucrose gradient reproducibly returned side bands corresponding to protein enriched (ρ ≈ 1.39 gcm−3) and protein depleted (ρ ≈ 1.42 gcm−3) chromatin fragments. We attribute this to the exchange and disproportionation of the linker H1 and H5 histones presumed to occur at the higher NaCl concentrations in the sucrose gradient.33,34 This linker histone exchange may also explain the reproducible lack of a sharp band in the sucrose gradient at 120 mM NaCl for the 16.17 Kbp β-globin chromatin fragment. As the molecular mass of the chromatin fragments remains essentially unchanged, the sedimentation data were used to determine the frictional coefficients f at 100 mM NaCl (Table 1) for both heterochromatin fragments. The frictional coefficients are smaller than those determined at 80 mM NaCl, reflecting the decreased flexibility of the rod like fiber. Furthermore, the sedimentation coefficient, and hence the frictional coefficient, for the 15.51 Kbp condensed chromatin fragment is essentially unchanged as the NaCl concentration is increased from 100 to 120 mM NaCl. The predicted dependencies of the frictional coefficients on rod length for rod diameters of 33 and 45 are shown in Figure 3D. These dependencies, based on the theory of Broersma,28,29 demonstrate that the experimental frictional coefficients are consistent with chromatin fibers having 6 or 7 nucleosomes and diameters of the order of 33 to 45 nm. The experimental data points for the 15.51 Kbp fragment at 80 and 120 mM NaCl are marked by double arrows (Figure 3D), whereas the ellipse marks the predicted value of f for the much shorter rods that would correspond to the structures seen by Robinson et al.22 The results indicate that no major structural changes have occurred as the NaCl concentration is raised, and that the hydrodynamic properties of the heterochromatic fragment are inconsistent with the formation of a much shorter and more compact rod.
Table 1.
Modeling of the frictional coefficients using the theory of Broersma.
| Heterochromatin fragment | HpaII, constitutive | NheI and XhoI, facultative |
|---|---|---|
| DNA size | 15.51 Kbp | 16.17 Kbp |
| f (80mM NaCl) | (1.07 ± 0.06) × 10−6 gs−1 a | (0.96 ± 0.02) × 10−6 gs−1 |
| f (100mM NaCl) | (0.99 ± 0.06) × 10−6 gs−1 | (0.93 ± 0.06) × 10−6 gs−1 |
| Rod-length at d = 33 nm b | 174 nm | 154 nm |
| Nucleosomes per turn | 5.6 | 6.2 |
| Rod-length at d = 45 nm b | 136 nm | 127 nm |
| Nucleosomes per turn | 7.2 | 7.5 |
Data from reference 19.
Modeling based on the frictional coefficients determined at 100 mM NaCl.
Discussion
In an earlier study we measured the hydrodynamic properties of a 15.51 Kbp constitutively condensed heterochromatin segment upstream of the chicken β-globin locus, and showed that its hydrodynamic behavior is similar to that expected for a rod like particle 33 – 45 nm in diameter having about 6 nucleosomes per 11 nm in length. To understand better the possible differences and similarities in the structure of constitutive and facultative heterochromatin, we compared the physical properties of this 15.51 Kbp constitutively condensed chromatin region with those of an adjacent 16.17 Kbp fragment containing portions of the β-globin locus control region and gene cluster. In the 6C2 cells used, the globin genes are transcribed only at very low levels. Using a combination of NgomIV, XhoI and NheI restriction endonucleases, both fragments were released allowing for a direct comparison of their sedimentation properties within the same sucrose gradient. The facultative heterochromatin 16.17 Kbp β-globin fragment sediments with a sedimentation coefficient of 138 ± 2 S, faster than the 15.51 Kbp constitutive heterochromatin fragment (s20,w = 128 ± 7 S).19 The protein content of the particle and its molecular weight were determined by CsCl density gradient banding of the cross-linked particle. Unlike the 15.51 Kbp constitutive heterochromatin fragment, which bands within a narrow density range, the 16.17 Kbp β-globin fragment show a broader density distribution centered on 1.424 gcm−3, similar to that observed for bulk chromatin.16,17 These observations reveal an overall decrease in the ratio of protein to DNA in the β-globin fragment as compared with the heterochromatin fragment, and also demonstrate the importance of the information obtained from CsCl gradients for analyzing individual chromatin fragments. Even though the molecular mass determined for the 16.17 Kbp β-globin chromatin fragment was slightly smaller than that observed for the 15.51 Kbp condensed chromatin fragment, showing that it has a lower protein:DNA ratio and lower nucleosome density, this did not result in a less compact structure. In fact, the frictional coefficient is consistent with rod like structures accommodating 6 – 7 nucleosomes per turn for diameters of 33 – 45 nm. The increased compaction resulted from the shorter rod lengths, illustrating the versatility of the 30 nm chromatin fiber in its ability to accommodate different linker lengths. Furthermore, this increased compaction is not necessarily inconsistent with the differential restriction enzyme accessibility observed for the 15.51 Kbp and 16.17 Kbp heterochromatin fragments. In the latter case, the increased average linker length and the proximity of BamHI restriction sites to the hypersensitive sites, may account for these differences. Unlike the 15.51 Kbp constitutive heterochromatin fragment, the 16.17 Kbp facultative heterochromatin fragment contains various hypersensitive sites (Figure 1) which could in principle perturb the overall chromatin structure. It has been shown by Wachtel35 that a short segment of free DNA appended to a compact chromatin structure would only result in a minimal increase of the frictional coefficient. If the HS3 and β/ε enhancer ends of the 16.17 Kbp fragment were protein free, the expected negative contribution to the sedimentation coefficient, is well within our experimental error and can therefore be ignored. The internal hypersensitive sites, HS2 and HS1, located ~ 2 and 5 Kbp from the HS3 end, span approximately 200 bp36 each. If these formed a segment of free, extended DNA, we would expect sedimentation coefficients considerably smaller than those we have measured for the compact 15.51 Kbp condensed chromatin fragment. As this is not the case, we conclude that these hypersensitive sites are incorporated within the higher order chromatin structure without disrupting it. We note that although the 15.51 and 16.17 Kbp fragments show similar sedimentation behavior at the low and high salt limits, they behave somewhat differently at intermediate salt concentrations (Figure 3B). This may reflect differences in local chromatin organization and stability between the two kinds of chromatin during the folding process that accompanies the increase in ionic strength.
Small angle neutron scattering studies on bulk chromatin averaging 20 Kbp in length reveal a constant mass per unit length of 6 – 7 nucleosomes per turn at ionic strengths above 75 mM NaCl.8 However, sedimentation velocity studies on similarly sized bulk chromatin show that the sedimentation coefficient increases monotonically with the ionic strength between 75 and 120 mM NaCl.10,12 This may be attributed to a decreasing flexibility of the chromatin fiber with increasing salt, as the parameters measured by neutron scattering are not influenced greatly by breaks or kinks on the fiber. Both these physical studies were carried out on bulk chromatin obtained by micrococcal nuclease digestion of nuclei. Contrasting results were obtained in recent electron microscopic studies of structures containing between 47 and 72 nucleosomes, formed by reconstitution of histones on a template of repeated DNA elements, each containing the 601 nucleosome positioning sequence. For repeat lengths between 177 and 207 bp the observed mass per unit length is about 10 nucleosomes per turn, at salt concentrations higher than those used in previous studies.22 In order to accommodate such an arrangement, the nucleosomes within the one-start helix are assumed to be interdigitated, resulting in a far more compact structure than expected for the canonical 30 nm fiber having 6 – 7 nucleosomes per turn.
We considered the possibility that the specific heterochromatin fragments that we have been studying might in fact adopt such compact structures at higher salt concentrations, and therefore measured their sedimentation properties as a function of the ionic strength. The sedimentation coefficients for these discrete chromatin fragments increased monotonically with salt concentration as has been previously observed for bulk chromatin fragments.10,12 While there is some increase in the values of the sedimentation coefficients at 100 and 120 mM NaCl, consistent with loss of flexibility, the corresponding frictional coefficients are nonetheless compatible with extended rod like structures having 6 – 7 nucleosomes per turn. Furthermore, there is no evidence of the kind of abrupt change in sedimentation properties with increasing ionic strength that would be expected if a transition had occurred from a 6 to a 10 nucleosome per turn structure. The physical properties of the naturally occurring heterochromatin fragments studied are therefore different from those reported for the reconstitutes of Robinson et al.22 The regularity of the DNA repeats and linker lengths, together with the additional regularity imposed by the presence of the strong 601 nucleosome positioning signals, may contribute to formation of the structures they observe.
Can heterochromatin adopt even more compact structures than those we have found in the β-globin locus? Earlier studies of mouse major and minor satellite heterochromatin, obtained by treatment of nuclei with micrococcal nuclease or restriction enzymes, showed that fragments of this chromatin sediment more rapidly than bulk chromatin.17 Based on the protein content calculated from buoyant density measurements in CsCl gradients, it was concluded that the unusual sedimentation properties arise largely from shape differences and correspond to structures that are more compact than bulk chromatin. In more recent studies18 chromatin obtained from a human lymphoblastoid cell line was separated on a sucrose gradient and a single fraction was further characterized based on its DNA size. The smaller DNA fragments, representing the most compact chromatin fibers, were enriched in centromeric, constitutive heterochromatin (C-bands) as well as low gene density chromatin (G-bands). These fragments had a DNA size of ~ 10 Kbp, whereas the bulk chromatin in the same gradient fraction had a size of ~20 Kbp. If the bulk chromatin has a protein to DNA content similar to that observed for the 16.17 Kbp β-globin chromatin fragment, then the theory of Broersma indicates sedimentation coefficients of 157 and 133 S, for 33 and 45 nm diameter chromatin fibers, respectively. A protein rich 10 Kbp heterochromatin fragment, having a CsCl buoyant density of 1.40 gcm−3 and identical sedimentation coefficients, would then have frictional coefficients consistent with a degree of compaction much higher than any we have observed so far.
Our results provide evidence that the constitutive and facultative heterochromatin fragments excised from the β-globin locus assume similar, extended rod like forms, despite the fact that the nucleosome density on the 16.17 Kbp globin locus fragment is considerably lower than that found on the adjacent constitutively condensed 15.51 Kbp region. This reflects the adaptability of chromatin structure in accommodating different DNA linker lengths. The methods described here can be applied to other domains, and should open the way to detailed studies of the physical properties of defined chromatin fragments such as the open high gene density fragments, as well as the highly compact centromeric heterochromatin described above.
Materials and Methods
Restriction enzyme digestion of 6C2 nuclei
Restriction enzyme digestion of 6C2 nuclei was carried out as described.19 Briefly, nuclei were isolated from 12 T150 dishes of 6C2, washed at least twice in NEBuffer #2 (50 mM NaCl, 10 mM Tris-HCl, 10 mM MgCl2, 1 mM dithiothreitol (pH 7.9 at 25°C)) and resuspended in NEBuffer #2 to an A260 of 100. The nuclei were digested with 5U/μl of the various restriction enzymes for 4 hours at 37°C. Reactions were stopped by adding 1/3 volume of 30 mM EDTA, 30 mM EGTA. Digests were carried out with HpaII, XhoI and NheI, as well as NgomIV, XhoI and NheI. The digested nuclei were pelleted in a microcentrifuge for 7 minutes at 1,200 g, resuspended in 1.0 ml Buffer A (80 mM KCl, 10 mM NaCl, 0.5 mM EDTA, 0.1 mM EGTA, 0.5 mM spermidine, 0.15 mM spermine, 15 mM 2-mercaptoethanol, 15 mM Tris-HCl (pH 8.0), 0.5% Brij® 58) and incubated for 10 minutes at 4°C. A ½ volume of 2 M urea was added and nuclei were incubated at 4°C for 2 hours. Nuclei were then dialyzed overnight at 4°C in DB-80 buffer (80 mM NaCl, 10 mM Tris-HCl (pH 7.4), 0.2 mM EDTA) and pelleted at 16,000 g for 10 minutes at 4°C. The supernatant containing the soluble chromatin was retained for further analysis. All solutions contain the following protease inhibitors: 0.75 μg/mL pepstatin A, 0.50 μg/mL leupeptin, 1.0 μg/mL aprotinin and 0.2 mM PMSF.
Fractionation of the 15.51 and 16.17 Kbp chromatin fragments
Gradients of 5–30% (w/v) sucrose in DB-n buffer were poured in SW40 14 × 95 mm centrifuge tubes with a BioComp Gradient Master 107 as described.19 DB buffers containing 10 mM Tris-HCl (pH 7.4), 0.2mM EDTA of various ionic strengths were used, containing 20 (DB-20), 40 (DB-40), 60 (DB-60), 80 (DB-80), 100 (DB-100) and 120 (DB-120) mM NaCl. 400 – 500 μL of digested chromatin were layered on the gradients and centrifuged in a SW40Ti rotor at 38,000 rpm and 4.0°C for 1.5 hours. After centrifugation, 500 μL fractionations were collected from the top. In order to identify the fraction(s) containing the chromatin fragments of interest, DNA obtained from each fraction was purified by phenol/chloroform extraction and ethanol precipitation. The purified DNA samples were analyzed by real-time PCR on an ABI 7900HT sequence detector using DNA primers and Taqman probes specific to the 15.51 Kbp region (numbers 13.192 and 17.763),23 or the 16.17 Kbp fragment (numbers 27.649 and 39.807).23 In DB-80 the 15.51 Kbp condensed chromatin fragment was reproducibly found in fraction 14, whereas the 16.17 β-globin chromatin fragment was reproducibly found in fraction 15.
Southern blot analysis
Analysis of the chromatin fragments released from 6C2 nuclei by NheI and XhoI digestion was performed by size separation of the DNA on a 0.8% agarose gel in TAE run at 100 volts for 3 to 3 ½ hours. DNA of interest was sized by Southern blot analysis. DNA was purified from the sucrose gradient fractions by phenol/chloroform extraction and ethanol precipitation, analyzed by gel electrophoresis and transferred to a GeneScreen membane as described.37 Membranes were probed with probe 410, a 632 bp BamHI and PstI fragment just upstream of the βh-globin gene, specific for the 16.17 Kbp fragment. Blots were analyzed on a GE Healthcare Typhoon 8600 phosphorimager.
Determination of the buoyant density of the 15.51 and 16.17 Kbp fragments
Sucrose gradient fractions containing the 15.51 Kbp HpaII chromatin fragment, or the 16.17 Kbp NheI and XhoI chromatin fragment from 6C2 nuclei were dialyzed into 80 mM NaCl, 10 mM triethanolamine-NaOH (pH 7.6), 0.2 mM EDTA and cross linked with 0.5% formaldehyde as described.17 Excess cross-linker was removed by dialysis into DB-80 buffer. Samples were made up to 1.0 mL and treated with an equal volume of CsCl in dialysis buffer having a concentration of 1.06 gmL−1, to a final density of approximately 1.40 gcm−3. 2.2 mL of this solution was centrifuged in a Beckman TLS-55 rotor at 20.0°C and 30,000 rpm for 96 hours. Gradients were fractionated from the top into 20 × 100 μL fractions. 30 μL of each fraction was used to determine the refractive index, and thus the density. The remaining 70 μL were treated as described16 to obtain free DNA for analysis by quantitative PCR using the appropriate primer set and Taqman probe.23
Acknowledgments
The authors thank members of the Felsenfeld laboratory for helpful discussions and a critical reading of the manuscript. This work was supported by the intramural research program of the NIH, National Institute of Diabetes and Digestive and Kidney Diseases.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Finch JT, Klug A. Solenoidal model for superstructure in chromatin. Proc Natl Acad Sci USA. 1976;73:1897–1901. doi: 10.1073/pnas.73.6.1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Thoma F, Koller Th, Klug A. Involvement of histone H1 in the organization of the nucleosome and of the salt-dependent superstructures of chromatin. J Cell Biol. 1979;83:403–427. doi: 10.1083/jcb.83.2.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Langmore JP, Schutt C. The higher order structure of chicken erythrocyte chromosomes in vivo. Nature. 1980;288:620–622. doi: 10.1038/288620a0. [DOI] [PubMed] [Google Scholar]
- 4.Widom J, Klug A. Structure of the 300Å chromatin filament: X-ray diffraction from oriented samples. Cell. 1985;43:207–213. doi: 10.1016/0092-8674(85)90025-x. [DOI] [PubMed] [Google Scholar]
- 5.Bordas J, Perez-Grau L, Koch MHJ, Vega MC, Nave C. The superstructure of chromatin and its condensation mechanism. I Synchrotron radiation X-ray scattering results. Eur Biophys J. 1986;13:157–173. doi: 10.1007/BF00542560. [DOI] [PubMed] [Google Scholar]
- 6.Lowary PT, Widom J. Higher-order structure of Saccharomyces cerevesiae chromatin. Proc Natl Acad Sci USA. 1989;86:8266–8270. doi: 10.1073/pnas.86.21.8266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Suau P, Bradbury EM, Baldwin JP. Higher-order structures of chromatin in solution. Eur J Biochem. 1979;97:593–602. doi: 10.1111/j.1432-1033.1979.tb13148.x. [DOI] [PubMed] [Google Scholar]
- 8.Gerchman SE, Ramakrishnan V. Chromatin higher-order structure studied by neutron scattering and scanning transmission electron microscopy. Proc Natl Acad Sci USA. 1985;84:7802–7806. doi: 10.1073/pnas.84.22.7802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Graziano V, Gerchman SE, Schneider DK, Ramakrishnan V. Histone H1 is located in the interior of the chromatin 30-nm filament. Nature. 1994;368:351–354. doi: 10.1038/368351a0. [DOI] [PubMed] [Google Scholar]
- 10.Butler PJG, Thomas JO. Changes in chromatin folding in solution. J Mol Biol. 1980;140:505–529. doi: 10.1016/0022-2836(80)90268-5. [DOI] [PubMed] [Google Scholar]
- 11.Pearson EC, Butler PJG, Thomas JO. Higher-order structure of nucleosome oligomers from short-repeat chromatin. EMBO J. 1983;2:1367–1372. doi: 10.1002/j.1460-2075.1983.tb01593.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Thomas JO, Butler PJG. Size-dependence of a stable higher-order structure of chromatin. J Mol Biol. 1980;144:89–93. doi: 10.1016/0022-2836(80)90215-6. [DOI] [PubMed] [Google Scholar]
- 13.Thomas JO, Rees C, Butler PJG. Salt-induced folding of sea urchin sperm chromatin. Eur J Biochemistry. 1986;154:343–348. doi: 10.1111/j.1432-1033.1986.tb09403.x. [DOI] [PubMed] [Google Scholar]
- 14.Kimura T, Mills FC, Allan J, Gould H. Selective unfolding of erythroid chromatin in the region of the active β-globin gene. Nature. 1983;306:709–712. doi: 10.1038/306709a0. [DOI] [PubMed] [Google Scholar]
- 15.Fisher EA, Felsenfeld G. Comparison of the folding of β-globin and ovoalbumin gene containing chromatin isolated from chicken oviduct erythrocytes. Biochemistry. 1986;25:8010–8016. doi: 10.1021/bi00372a033. [DOI] [PubMed] [Google Scholar]
- 16.Caplan A, Kimura T, Gould H, Allan J. Perturbation of chromatin structure in the region of the adult β-globin gene in chicken erythrocyte chromatin. J Mol Biol. 1987;193:57–69. doi: 10.1016/0022-2836(87)90626-7. [DOI] [PubMed] [Google Scholar]
- 17.Gilbert N, Allan J. Distinctive higher-order chromatin structure at mammalian centromeres. Proc Natl Acad Sci USA. 2001;98:11949–11954. doi: 10.1073/pnas.211322798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gilbert N, Boyle S, Fiegler H, Woodfine K, Bickmore WA. Chromatin architecture of the human genome: gene-rich domains are enriched in open chromatin fibers. Cell. 2004;118:555–566. doi: 10.1016/j.cell.2004.08.011. [DOI] [PubMed] [Google Scholar]
- 19.Ghirlando R, Litt DM, Prioleau MN, Recillas-Targa F, Felsenfeld G. Physical properties of a genomic condensed chromatin fragment. J Mol Biol. 2004;336:597–605. doi: 10.1016/j.jmb.2003.12.049. [DOI] [PubMed] [Google Scholar]
- 20.Schlach T, Duda S, Sargent DF, Richmond TJ. X-ray structure of a tetranucleosome and its implications for the chromatin fibre. Nature. 2005;436:138–141. doi: 10.1038/nature03686. [DOI] [PubMed] [Google Scholar]
- 21.Dorigo B, Schalch T, Kulangara A, Duda S, Schroeder RR, Richmond TJ. Nucleosome arrays reveal the two-start organization of the chromatin fiber. Science. 2004;306:1571–1573. doi: 10.1126/science.1103124. [DOI] [PubMed] [Google Scholar]
- 22.Robinson PJJ, Fairall L, Huynh VAT, Rhodes D. EM measurements define the dimensions of the “30-nm” fiber: Evidence for a compact, interdigitated structure. Proc Natl Acad Sci USA. 2006;103:6506–6511. doi: 10.1073/pnas.0601212103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Litt MD, Simpson M, Recillas-Targa F, Prioleau MN, Felsenfeld G. Transitions in histone acetylation reveal boundaries of three separately regulated neighboring loci. EMBO J. 2001;20:2224–2235. doi: 10.1093/emboj/20.9.2224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Litt MD, Simpson M, Gaszner M, Allis CD, Felsenfeld G. Correlation between histone lysine methylation and developmental changes at the chicken β-globin locus. Science. 2001;293:2453–2455. doi: 10.1126/science.1064413. [DOI] [PubMed] [Google Scholar]
- 25.Boyes J, Felsenfeld G. Tissue-specific factors additively increase the probability of the all-or-none formation of a hypersensitive site. EMBO J. 1996;15:2496–2507. [PMC free article] [PubMed] [Google Scholar]
- 26.Prioleau MN, Nony P, Simpson M, Felsenfeld G. An insulator element and condensed chromatin region separate the chicken β-globin locus from an independently regulated erythroid-specific folate receptor gene. EMBO J. 1999;18:4035–4048. doi: 10.1093/emboj/18.14.4035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ausio J, Borochov N, Seger D, Eisenberg H. Interaction of chromatin with NaCl and MgCl2. Solubility and binding studies, transition to and characterization of the higher-order structure. J Mol Biol. 1984;177:373–398. doi: 10.1016/0022-2836(84)90291-2. [DOI] [PubMed] [Google Scholar]
- 28.Broersma S. Viscous force constant for a closed cylinder. J Chem Phys. 1960;32:1632–1635. [Google Scholar]
- 29.Newman J, Swinney HL, Day LA. Hydrodynamic properties and structure of fd virus. J Mol Biol. 1977;116:593–606. doi: 10.1016/0022-2836(77)90086-9. [DOI] [PubMed] [Google Scholar]
- 30.Minie M, Clark D, Trainor C, Evans T, Reitman M, Hannon R, Gould H, Felsenfeld G. Developmental regulation of globin gene expression. J Cell Sci. 1992;(Suppl 16):15–20. doi: 10.1242/jcs.1992.supplement_16.3. [DOI] [PubMed] [Google Scholar]
- 31.Thåström A, Lowary PT, Widlund HR, Cao H, Kubista M, Widom J. Sequence motifs and free energies of selected natural and non-natural nucleosome positioning sequences. J Mol Biol. 1999;288:213–229. doi: 10.1006/jmbi.1999.2686. [DOI] [PubMed] [Google Scholar]
- 32.Huynh VAT, Robinson PJJ, Rhodes D. A method for the in vitro reconstitution of a defined “30 nm” chromatin fibre containing stoichiometric amounts of the linker histone. J Mol Biol. 2005;345:957–968. doi: 10.1016/j.jmb.2004.10.075. [DOI] [PubMed] [Google Scholar]
- 33.Thomas JO, Rees C. Exchange of histones H1 and H5 between chromatin fragments. A preference of H5 for higher-order structures. Eur J Biochem. 1983;134:109–115. doi: 10.1111/j.1432-1033.1983.tb07538.x. [DOI] [PubMed] [Google Scholar]
- 34.Komaiko W, Felsenfeld G. Solubility and structure of domains of chicken erythrocyte chromatin containing transcriptionally competent and inactive genes. Biochemistry. 1985;24:1186–1193. doi: 10.1021/bi00326a020. [DOI] [PubMed] [Google Scholar]
- 35.Wachtel EJ. Calculation of hydrodynamic properties of model chromatin structures containing unfolded regions. J Mol Biol. 1987;193:69–70. doi: 10.1016/0022-2836(87)90627-9. Appendix to reference 16. [DOI] [PubMed] [Google Scholar]
- 36.Abruzzo LV, Reitman M. Enhancer activity of upstream hypersensitive site 2 of the chicken β-globin cluster is mediated by GATA sites. J Biol Chem. 1994;269:32565–32571. [PubMed] [Google Scholar]
- 37.Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning, A Laboratory Manual. Cold Spring Harbor Laboratory Press; Cold Spring Harbor: 1989. pp. 9.42–9.58. [Google Scholar]