Abstract
Epstein Barr virus (EBV) is associated with B cell lymphomas in post-transplant lymphoproliferative disease (PTLD). Latent membrane protein 1 (LMP1), the major oncogenic protein of EBV, promotes tumorigenesis through activation of NF-κB, Erk, p38, JNK, and Akt. The Jak/STAT signal transduction pathway is also constitutively active in PTLD-associated EBV+ B cell lymphomas. Here we determine the mechanism of Jak/STAT activation in EBV+ B cell lymphomas and the role of LMP1 in this process. Immunoprecipitation studies revealed no direct interaction of LMP1 and JAK3, but known associations between JAK3 and common gamma chain, and between LMP1 and TRAF3, were readily detected in EBV+ B cell lines from patients with PTLD. An inducible LMP1 molecule expressed in EBV- BL41 Burkitt’s cells demonstrated STAT activation only after prolonged LMP1 signaling. While LMP1 induced IFN-γ production in BL41 cells, IFN-γ receptor blockade and IFN-γ neutralization prior to LMP1 activation markedly decreased STAT1 activation and expression of LMP1-driven IFN-γ inducible genes. Understanding the mechanisms by which EBV induces cellular signal transduction pathways may facilitate development of new treatments for PTLD.
Introduction
PTLD is a serious, and potentially life-threatening, complication of solid organ transplantation. The majority of PTLD are B cell lymphoproliferations associated with EBV. In the most severe form, PTLD manifests as aggressive, malignant EBV+ B cell lymphoma. LMP1 is an EBV-encoded protein that is required for the establishment and continued proliferation of human B lymphoblastoid cell lines (LCL) and is sufficient to induce transformation of rodent fibroblasts (1, 2). Further, LMP1 is expressed on the membrane of PTLD-associated B cell lymphomas and mimics the function of activated CD40. LMP1 is constitutively active, in the absence of any known ligand, when expressed in the cell membrane. The six transmembrane spanning domains of LMP1 induce clustering of LMP1 complexes that facilitate formation of docking sites for cellular signaling adaptor proteins and results in constitutive activation. LMP1 initiates several cellular signaling pathways including NF-κB, PI3K/Akt and the mitogen-activated protein (MAP) kinases ERK, JNK, and p38 that are essential for growth and survival of transformed B cells (3-7).
LMP1 has also been reported to directly activate the Jak/STAT pathway, critical for cytokine receptor signaling, although these results are controversial (8, 9). Jak family members constitutively associate with cytokine receptors and upon cytokine binding to the receptor become autophosphorylated. Jak also phosphorylate the cytokine receptor on tyrosine (Tyr) residues that serve as docking sites for cytoplasmic STAT proteins. Finally, Jak phosphorylate receptor-associated STAT proteins permitting STAT dimer formation and localization to the nucleus where STAT homodimers or heterodimers bind to promoter sequences of STAT-inducible genes to regulate their transcription (10-13).
In normal cells, STAT activity is highly regulated and activation is transient. However, many EBV-related malignancies, including PTLD, are characterized by dysregulation of the Jak/STAT pathway. We reported that STAT1 and STAT3, and associated Jak, are constitutively phosphorylated at Tyr residues in EBV+ spontaneous LCL (SLCL) from PTLD patients. We also demonstrated that STAT3 is constitutively active in primary PTLD tumors and in EBV+ B cell lymphomas that arise in SCID mice injected with SLCL (14, 15).
The factors governing STAT activation in EBV+ B cell lymphomas remain unclear. Previous studies by Najjar et al (16), using an in vitro generated EBV+ lymphoblastoid cell line (LCL), suggested that constitutive STAT1 activation results from NF-κB-dependent-secretion of interferons. The aim of the current study was establish whether a similar pathway was active in PTLD-associated B cell lymphomas, to evaluate the specific role of LMP1 in STAT activation, and to determine the downstream significance of STAT1 activation in PTLD-associated B cell lymphomas.
Materials and Methods
Reagents and Cell Lines
Anti-human nerve growth factor receptor (NGFR) was from Sigma-Aldrich (St. Louis, MO) and biotinylated anti-NGFR from Chromaprobe (Maryland Heights, MO). Goat anti-mouse IgG, HRP-conjugated goat anti-rabbit, and HRP-conjugated donkey anti-mouse Abs were purchased from Jackson ImmunoResearch Laboratories. Streptavidin-PE was from BD Pharmingen (San Jose, CA). For immunoprecipitation (IP) and western blotting the following antibodies were used: anti-STAT1α (C-24), anti-STAT3, anti-pTyrSTAT3, anti-common γ chain (Santa Cruz Biotechnology, Santa Cruz, CA), anti-pTyr701 STAT1 (Cell Signaling Technologies, Boston, MA), anti-LMP1 (CS1-4) (Dako, Denmark) and anti-β actin (Sigma-Aldrich). LY294002, SB203580, and Bay 11-7082 were from Calbiochem (San Diego, CA). The BL41 Burkitt’s lymphoma cell line and BJAB B cell lymphoma line were gifts from E. Kieff (Harvard Medical School, Boston, MA). AB5, JB7, MF4, and VB5 are spontaneously generated EBV+ LCL from peripheral blood or lymph nodes of PTLD patients (14), termed SLCL. Cells were grown in RPMI 1640 , 10% FCS (Mediatech, Manassas, VA) and Penicillin-Streptomycin (Invitrogen Life Technologies, Carlsbad, CA).
Immunoprecipitation and Western Blotting
1.5×108 cells were homogenized in 0.5% Brij58 buffer (0 mM Tris pH7.4, 100 mM NaCl, 1 mM EDTA, 1 mM PMSF, 20 μg/ml of aprotinin) and lysates cleared by centrifugation. The lysate was pre-cleared with protein G beads (Amersham Pharmacia, Pittsburgh, PA) and immunoprecipitated with anti-LMP1, anti-γ chain, or IgG antibodies. Immune complexes were pulled down with protein G beads and resolved on 7.5% SDS-PAGE, transferred to nitrocellulose membranes and Weestern blotting performed. For STAT IPs, 5×106 cells were washed with cold PBS/1 mM orthovanadate and lysed in 1% NP40/0.5% deoxycholic acid phospholysis buffer (150 mM NaCl, 0.5 mM EDTA, 10 mM NaF, 2 mM PMSF, 50 μg/ml aprotinin, 50 μg/ml leupeptin, 5 μg/ml pepstatin). Lysates were cleared by centrifugation, incubated with anti-STAT1 or anti-STAT3 antibodies, complexes pulled down by protein G beads and analyzed by Western blotting with anti-pTyrSTAT1, anti-STAT1, anti-pTyrSTAT3, and anti-STAT3 antibodies.
Generation of BL41 Clones Expressing NGFR-LMP1
NGFR-LMP1 chimeras were created as described previously (17). LMP1 was isolated from SLCL to generate tumor-derived NGFR-LMP1. The C-terminus of LMP1 was cloned from SLCL with the same primers used for isolation of LMP1 from the B95.8 strain of EBV. NGFR was cloned from the vector and joined to the C-terminus of LMP1 by SOEing PCR. The product was introduced into pcDNA3. BL41 cells were electroporated at 210V, 960 μF with 15 μg of pcDNA3.NGFR-LMP1 construct. Cells were plated in RPMI with 0.7 mg/ml of G418 (Invitrogen Life Technologies). Selected clones were expanded and screened for NGFR-LMP1 expression by immunofluorescent staining and flow cytometry.
Flow Cytometry
Cells were labeled with biotinylated anti-NGFR mAb followed by streptavidin-PE or with anti-IFNR1-PE, anti-IFNRII-PE and analyzed by flow cytometry using a Becton Dickinson FACscan.
Electromobility Shift Assays
BL41.NGFR-LMP1 clones were incubated with anti-NGFR mAb for 30 minutes followed by goat anti-mouse IgG for 30 minutes or for 24 hours with or without pre-treatment with 40 μM brefeldin A. Cells were lysed in buffer (20 mM HEPES, pH 7.0, 300 mM NaCl, 10 mM KCl, 1 mM MgCl2, 0.1% Triton X-100, 0.5 mM dithiothreitol (DTT), 2 mM PMSF, 10 mM NaF, 1 mM Na3VO4, 5 μg/mL aprotinin, 5 μg/mL leupeptin, 0.5 μg/mL pepstatin, 20% glycerol). For each reaction, 40 μg of lysate and a double-stranded high-affinity serum-inducible element (hSIE) probe (Santa Cruz Biotechnology) end-labeled with [32P]ATP (PerkinElmer, Boston, MA) were incubated in binding buffer (12 mM HEPES, pH 7.9, 50 mM KCl, 5 mM MgCl2, 0.12 mM EDTA, 0.06 mM EGTA, 10% glycerol, 1 mM DTT, 0.1 mg/mL salmon sperm DNA (Life Technologies, Gaithersburg, MD), 0.1 mg/mL poly(dI-dC) (Amersham Pharmacia Biotech, Pittsburgh, PA)), with or without a 100-fold molar excess of unlabeled probe. Complexes were electrophoresed on 4% nondenaturing polyacrylamide gels and exposed to film for 24-48 hours at -70°C.
Luminex Assay and IFN-γ ELISA
BL41.NGFR-LMP1 clones were incubated with anti-NGFR mAb and goat anti-mouse IgG or with goat anti-mouse IgG alone for 24 hours. Supernatants were collected, pre-cleared with protein G beads, assayed in the Upstate Beadlyte 22-plex human cytokine assay (Millipore, Billerica, MA) according to the manufacturer’s protocol and analyzed on a Luminex 200 system. Supernatants were also analyzed in the High Sensitivity human IFN-γ ELISA (Abcam, Cambridge MA) according to the manufacturer’s protocol.
Quantitative PCR
BL41.NGFR-LMP1 clones were cross-linked for 0 or 24 hours with or without pre-incubation with anti-IFNR1 (2 μg/ml) and anti-IFNγ (20 μg/ml) antibodies or mouse IgG (22 μg/ml). Total RNA was isolated using TRIzol (Invitrogen Life Technologies) and first strand cDNA generated with oligo-dT primers (Invitrogen Life Technologies). Quantitative real-time PCR was performed on a StratageneMx3000P using SYBR Green PCR master mix (Applied Biosystems, Foster City, CA). The primers used were: 5′-GCCCGTGGACAGCGAGCAG3′ and 5′-GCCGGCGTTTGGAGTGGTAGA-3′ for p21; 5′-TGCAAGGAACCCCAGTAGTGA-3′ and 5′-GGTGGATAGTCCCTTGGTTGG-3′ for Mig1; 5′ACCACAGTCCATGCCATCAC-3′ and 5′-TCCACCACCCTGTTGCTGTA-3′ for GAPDH.
Statistics
Students t-test analysis was performed on gene expression levels of p21 and Mig1. p values ≤ 0.05 were considered statistically significant.
Results
LMP1 activates STAT without direct JAK3 interaction in PTLD-derived EBV+B cell lines
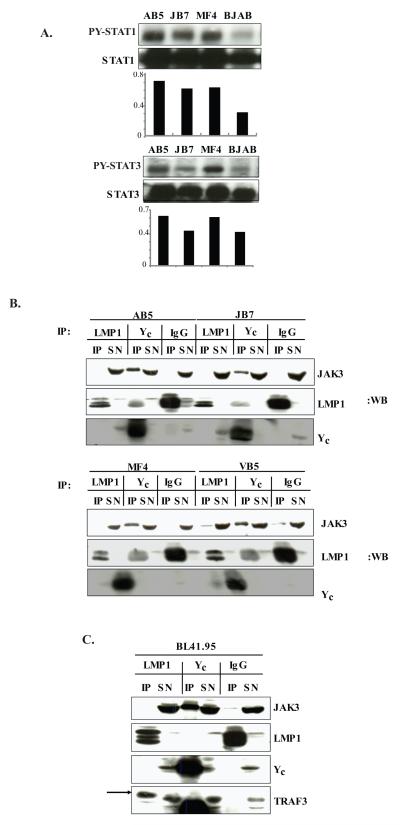
PTLD-derived SLCL AB5, JB7, MF4 and the EBV- B cell lymphoma BJAB were analyzed for tyrosine phosphorylation (p-Tyr) of STAT by immunoprecipitation with STAT1 or STAT3 antibodies followed by phospho-specific anti-STAT western blotting. All SLCL have constitutive p-Tyr STAT1 while the BJAB cell line shows minimal-low p-TyrSTAT1. Two of three SLCL have elevated p-Tyr STAT3, while one SLCL and the EBV- BJAB line show moderate p-Tyr STAT3 (Fig. 1A).
Figure 1. Activation of STAT1 and STAT3 in SLCL does not occur through LMP1 interaction with JAK3.
A. The SLCL, AB5, JB7, MF4 (1×107) or BJAB cells were lysed in phospholysis buffer and immunoprecipitated with either anti-STAT1 or anti-STAT3 antibodies. The immunoprecipitated proteins (IP) or post-immunoprecipitation supernatants (SN) were resolved on 7.5% SDS-PAGE and Western blotted with anti-phospho-Tyr STAT1, anti-phospho-Tyr STAT3, anti-STAT1 or anti-STAT3 antibodies. Densitometry results are shown below the blot. B. The SLCL AB5, JB7, MF4 and VB5 (1×108 cells) were lysed and immunoprecipitated with anti-LMP1, anti-γc or IgG antibodies. The proteins were resolved on 10% SDS-PAGE, transferred to nitrocellulose and analyzed for interaction with JAK3 by Western blotting with anti-JAK3 antibody. Additionally blots were incubated with anti-LMP1 and anti- γc antibodies. C. BL41_B95 were lysed and immunoprecipitated with anti-LMP1, anti-γc or IgG antibodies and analyzed for interaction with JAK3 and TRAF3 by Western blotting with appropriate antibodies as described above. The results are representative of three independent experiments.
To test whether JAK3 interacts with LMP1 to directly initiate STAT signaling in EBV+ cell lines from patients with PTLD, lysates from the AB5, JB7, MF4, and VB5 cell lines were immunoprecipitated with anti-LMP1 antibodies, anti-common γ chain (γc) antibodies as a positive control for known interaction of JAK3 with a transmembrane receptor, or IgG as a negative control. No specific interaction between LMP1 and JAK3 was observed in any of the cell lines although JAK3 did co-immunoprecipitate with γc as expected (Fig.1B). Further, JAK3 protein could be detected in immunoblots of post-immunoprecipitation supernatants (SN) indicating JAK3 was present in cell lysates. Background bands in LMP1 blots of γc immunoprecipitations were IgG heavy chain that runs at approximately the same size as LMP1. The IgG heavy chain band is also present as a second band in the LMP1 immunoprecipitations (LMP1 IP lanes). To determine whether differences in the origin of LMP1 accounted for the lack of LMP1/JAK3 interaction in EBV+ B cell lines from PTLD patients, we examined the association of LMP1 and JAK3 in the BL41_B95.8 B lymphoma cell line. BL41_B.95 was generated by infecting the EBV- parental BL41 line with the B95.8 EBV strain in vitro and, thus, expresses the B95.8 form of LMP1. No LMP1-JAK3 interaction was observed in BL41_B95 cells (Fig.1C). In contrast, LMP1 did co-immunoprecipitate with TRAF3, demonstrating the known LMP1/TRAF interaction was intact. Thus, JAK3 does not directly interact with either LMP1 from the B95.8 strain of EBV, or with LMP1 in EBV-infected PTLD-derived cell lines.
Activation of STAT1 by LMP1 is delayed
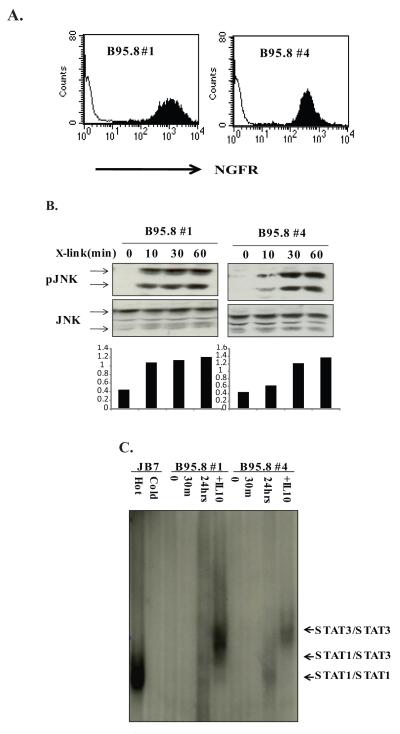
BL41 cells expressing NGFR-LMP1 chimeric proteins composed of the extracellular and transmembrane domains of NGFR and the C-terminus of the B95.8 variant of LMP1 were constructed (17). Cell surface expression of NGFR-LMP1 was confirmed by staining for NGFR and analyzed by flow cytometry. Two clones (B95.8 #1 and B95.8#4) were selected for further use (Fig.2A). NGFR-LMP1 crosslinking rapidly induced JNK phosphorylation in these clones indicating LMP1 signaling is intact (Fig. 2B).
Figure 2. LMP1 induces STAT1/3/DNA complexes 24 hr after activation.
A. BL41 clones expressing chimeric NGFR-LMP1 B95.8 were created as described in Materials and Methods and are referred to as B95.8 #1 and B95.8 #4. Expression of NGFR-LMP1 on the surface of BL41 cells was tested by flow cytometry using anti-NGFR-biotinylated antibodies and streptavidin-PE. Filled histogram shows NGFR staining in BL41 clones and thin line histogram indicates NGFR background staining in the untransfected parental BL41 line. B. B95.8 #1 and B95.8 #4 clones were incubated with anti-NGFR and goat anti mouse IgG for 0, 10, 30 or 60 minutes. The cells were harvested, lysed in phospholysis buffer and resolved on SDS-PAGE. After transfer to nitrocellulose, anti-phospho-JNK and anti-JNK antibodies were used for Western blot. Densitometry analysis is shown below the blots and represents the ratio of pJNK to total JNK. C. NGFR-LMP1 BL41 clones were crosslinked with anti-NGFR and goat anti-mouse Ig for 0, 30 minutes or 24 hours or treated with 10 ng/ml of recombinant human IL-10 for 30 minutes. 40 μg of whole cell lysates of BL41 clones and JB7 SLCL were incubated with 32P labelled hSIE probe with/without 1000x excess of cold probe and resolved on 4% non-denaturing acrylamide gel followed by autoradiography. Data are representative of three or more independent experiments.
Activation of STAT by LMP1 was analyzed by EMSA using a double-stranded 32P labeled hSIE probe containing a consensus-binding site for both STAT1 and STAT3. As expected, STAT/DNA complexes were evident in JB7 cells and could be competed away with excess cold probe (Fig. 2C). NGFR-LMP1 cross-linking induced STAT/DNA complexes at 24 hours, but not at 30 minutes, in both the BL41_B95.8 clone #1 and the BL41_B95.8 clone #4 (Fig. 2C). Analysis at 10 and 20 minutes also showed no STAT activation (data not shown). Thus, initiation of LMP1 signaling results in delayed activation of STAT. Mobility of the STAT/DNA complexes indicates that LMP1 signaling in BL41 clones primarily induced STAT1 homodimers and some STAT1/STAT3 heterodimers, whereas addition of exogenous IL-10 mainly induced STAT3 homodimers (Fig. 2C).
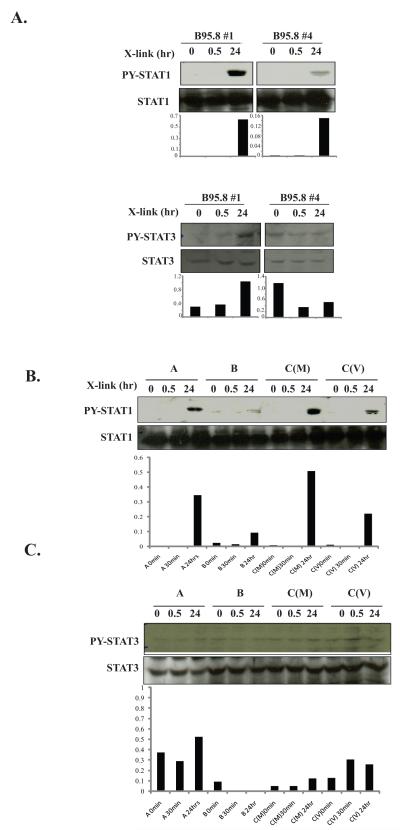
To further determine which STAT were activated by LMP1, NGFR-LMP1 BL41 clones were stimulated by NGFR cross-linking for 30 minutes or 24 hours and immunoprecipitation of STAT1 and STAT3 were performed. Both clone B95.8#1 and clone B95.8#4 induced p-Tyr of STAT1 24 hours after NGFR cross-linking (Fig. 3A, top) but only clone B95.8#1, which has the highest expression level of NGFR-LMP1 (Fig. 2A), also induced p-Tyr-STAT3 (Fig. 3A, bottom).
Figure 3. Tyrosine phosphorylation of STAT1 by LMP1 is delayed.
A. NGFR-LMP1 in B95.8#1 and B95.8#4 was crosslinked for 0, 30 minutes or 24 hours, cells harvested, and lysed in phospholysis buffer. Immunoprecipitation with anti-STAT1 antibody was performed followed by anti-p-Tyr- STAT1 and anti-STAT1 Western blot (left panels). For STAT3, 60 μg of lysates were resolved on SDS-PAGE and western blotted with anti-p-Tyr- STAT3 or anti-STAT3 antibodies (right panels). Densitometry analysis is shown below the blots and represents the ratio of p-Tyr STAT to total STAT. B. BL41 clones expressing NGFR-LMP1 from SLCL were incubated with anti-NGFR/goat anti-mouse antibodies for 0, 30 minutes or 24 hours and lysed. STAT1 was immunoprecipitated as in A. followed by Western blotting with anti-p-Tyr STAT1 and anti-STAT1 antibodies. Densitometry analysis is shown below the blots and represents the ratio of p-Tyr-STAT1 to total STAT1. C. BL41 tumor variant clones were treated as in B. and 60 μg of lysate was resolved on SDS-PAGE followed by Western blotting with anti-phospho-Tyr-STAT3 and anti-STAT3 antibodies. Densitrometry analysis is shown below the blots and represents the ratio of p-Tyr-STAT3 to total STAT3. Data for each set is representative of three independent experiments.
To establish whether delayed activation of STAT is also observed with variants of LMP1 other than the B95.8 form, BL41 clones expressing NGFR-LMP1 containing the C-terminus of LMP1 isolated from PTLD-derived SLCL were studied. The clones were named according to the LMP1 group nomenclature (18) but group C clones were derived from two different cell lines, MF4 and VB5, obtained from two different PTLD patients and are indicated as C(M) and C(V) respectively. STAT1 is only activated after 24 hours of NGFR-LMP1 cross-linking, while no significant increase in p-Tyr STAT3 was detected at either 30 minutes or 24 hours (Fig 3B, 3C). These results indicate that LMP1 derived from both the B95.8 strain of EBV, and from EBV+ PTLD cell lines, primarily induce the activation of STAT1 and that this activation is delayed.
LMP1 signaling induces a secreted factor that is responsible for p-Tyr STAT1
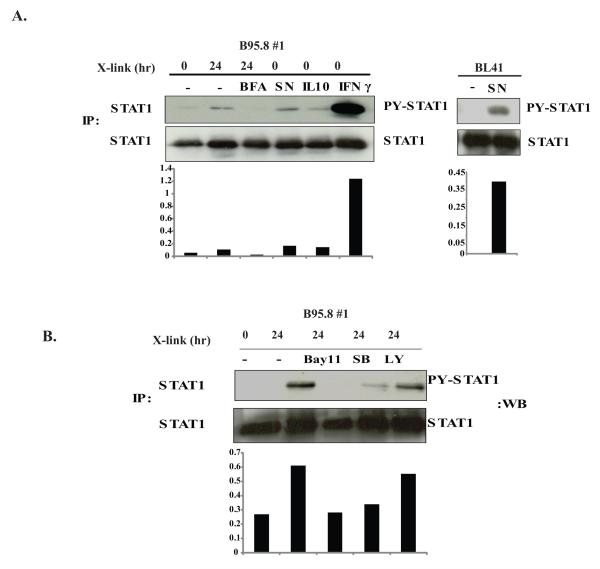
We next investigated whether activation of STAT by LMP1 occurs through an indirect pathway involving a secreted factor that induces p-TyrSTAT1 as had previously been suggested by the work of Najjar et al (16). We first tested whether inhibition of protein secretion can prevent p-TyrSTAT1 induced by NGFR-LMP1 crosslinking. Indeed, pre-incubation with the protein transport inhibitor BFA prevented p-TyrSTAT1 at 24 hours (Fig 4A lane 3, top left panel). Supernatants obtained from BL41.NGFR-LMP1 clones after NGFR cross-linking induced p-Tyr STAT1 in unstimulated BL41.NGFR-LMP1 cells (Fig. 4A lane 4, top left panel) as well as in non-transfected, non-stimulated, parental BL41 cells (Fig 4A, top right panel). Together, these data suggest that STAT1 activation occurs downstream of a receptor that binds a secreted protein induced by LMP1 signaling.
Figure 4. LMP1 contributes to phosphorylation of STAT1 through induction of a secreted NF-κB-dependent factor.
A, left panel. NGFR-LMP1 expressed in B95.8#1 was crosslinked for 0 hours or 24 hours without (lane 1,2) or with pre-incubation with Brefeldin A (lane 3). Supernatant from NGFR-LMP1-expressing cells crosslinked for 24 hours (lane 4) or exogenous IL-10 (lane 5) or exogenous INF-γ (lane 6) were added to uncrosslinked B95.8#1 for 30 minutes. All cells were lysed in phospholysis buffer and STAT1 was immunoprecipitated using anti-STAT1 antibody. The proteins were resolved on SDS-PAGE and Western blotted with anti-pTyr-STAT1 and anti-STAT1 antibodies. Densitometry analysis is shown below the blot and represents the ratio of p-Tyr STAT1 to total STAT1. A, right panel. Parental BL41 cells were cultured without (-) or with (SN) supernatant from NGFR-LMP1-expressing cells crosslinked for 24 hours and analyzed for phosphorylation of STAT1 as described above. B. B95.8 #1 cells were pre-incubated with either Bay11-7082 (1 μM), SB203580 (5 μM) or LY294002 (5 μM) for 30 minutes before crosslinking of NGFR-LMP1 for 24 hours. Lysates were prepared and STAT1 was immunoprecipitated followed by phospho-STAT1 and STAT1 Western blot. Densitometry analysis is shown below the blot and represents the ratio of p-Tyr-STAT1 to total STAT1. Data is representative of 3 independent experiments.
STAT1 is activated downstream of multiple cytokine receptors including IL-10R and IFN-γR. IL-10 can be induced by LMP1 (17), is an autocrine growth factor for SLCL (19), and therefore, was a candidate for mediating the indirect activation of STAT1 after LMP1 signaling. However, addition of high levels of exogenous IL-10 (10 ng/ml) to non-crosslinked BL41.NGFR-LMP1 clones induced only modest levels of p-TyrSTAT1 while low doses of IFN-γ (1 U/ml) were sufficient to induce very high levels of p-TyrSTAT1 (Fig.4A lanes 5, 6). Whereas IFN-γ induction is regulated by NF-κB and p38 but is independent of PI3K (20), IL-10 induction by LMP1 relies on p38 and PI3K (17). Therefore, BL41.NGFR-LMP1 clones were pre-incubated with the NF-κB inhibitor Bay11-7082, the p38 inhibitor SB203580 or the PI3K inhibitor LY294002, prior to NGFR cross-linking. STAT1 phosphorylation was abolished by Bay11-7082 and significantly decreased by SB203580, although LY294002 had minimal effect (Fig. 4B). These results suggest that phosphorylation of STAT1 through LMP1 signaling requires NF-κB and p38 activation, but not PI3K activation and support the possibility that IFN-γ, but not IL-10, is primarily responsible for indirect STAT1 phosphorylation.
LMP-induced production of IFN-γ leads to p-TyrSTAT1 and STAT1-inducible gene expression
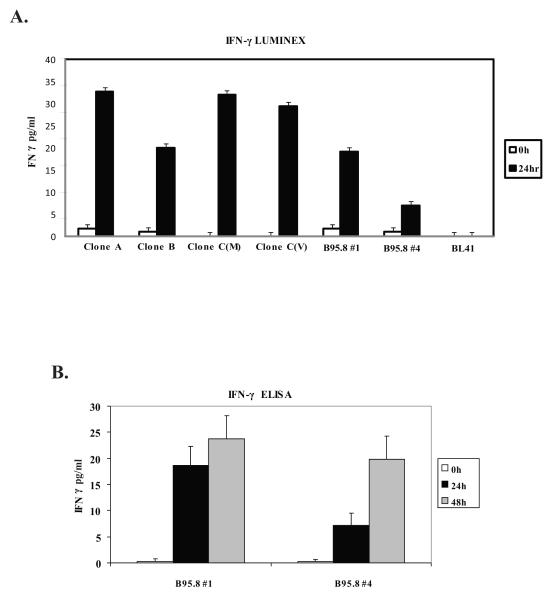
To investigate the LMP1-induced proteins responsible for STAT1 activation, BL41 clones expressing either NGFR-LMP1 B95.8 or NGFR-LMP1 derived from the PTLD tumor cell lines, were cultured in the absence of cross-linking, or cross-linked for 24 hours, and supernatants collected for the luminex assay. Cytokines induced by B95.8-derived LMP1, and LMP1 isolated from EBV+ SLCL, were similar and included IL-12p40, IL-15, IL-1α, IL-8, and TNF-α as well as IL-2 and IFN-γ, both known to activate STAT1 (data not shown). IFN-γ was induced after LMP1 signaling as detected in supernatants of NGFR-LMP1-BL441 cells by the luminex assay (Fig. 5A) and by ELISA (Fig. 5B) but was not detected in supernatants from the NGFR-LMP1 negative parental BL41 cell line (Fig 5A). Further, recombinant IFN-γ induced p-TyrSTAT1 in BL41.NGFR-LMP1 cells in the absence of NGFR crosslinking (Fig.4A, lane 6).
Figure 5. LMP1 Induces IFN-γ production and secretion by BL41 clones.
NGFR-LMP1 expressing BL41 clones containing either the tumor variant (A, B, C(M), C(V)) or the B95.8 isoform of LMP1 were crosslinked for 0 hours (white bars), 24 hours (black bars) or 48 hours (gray bars). Supernatants were harvested, pre-cleared with protein G beads and used in either A) pan cytokine luminex assay or B) high sensitivity IFN-γ ELISA. Results are shown as the mean pg/ml of IFN-γ per million cells. For luminex data, one of three independent experiments is shown. For ELISA, the data is a combination of two independent experiments.
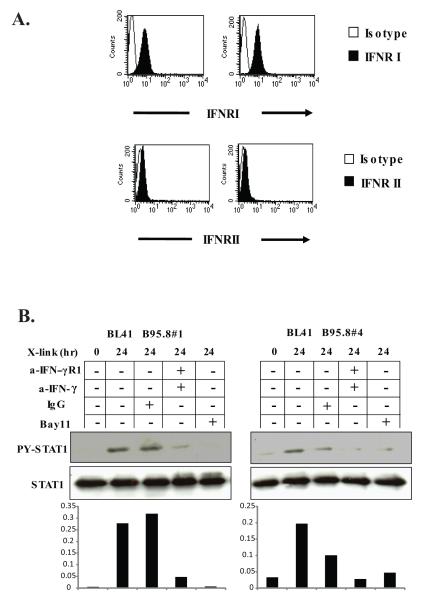
Next we tested whether the subunits of the IFN-γ receptor, IFNRI and IFNRII, were present on the surface of BL41.NGFR-LMP1 clones. Labeling with anti-IFNRI-PE and anti-IFNRII-PE antibodies and flow cytometry demonstrated that both subunits of the receptor are expressed by BL41 clones indicating that the receptor is available for IFN-γ binding and downstream signaling (Fig.6A). To determine whether IFN-γ is the key cytokine governing the indirect activation of STAT1 by LMP1, BL41.NGFR-LMP1 B95.8 cells were pre-incubated with blocking anti-IFNRI and neutralizing anti-IFN-γ antibodies or control IgG prior to cross-linking of NGFR-LMP1. Blockade of the IFN-γ receptor and neutralization of IFN-γ, or inhibition of NF-κB activation with Bay11, significantly decreased the levels of p-TyrSTAT1 in both BL41.NGFR-LMP1 clones tested (Fig. 6B). In contrast, pre-incubation with control IgG prior to NGFR crosslinking, had minimal, if any, effect on p-Tyr STAT1. Similarly, antibodies to IL-10 did not affect p-TyrSTAT1 when NGFR was crosslinked (data not shown).
Figure 6. IFN-γ participates in the indirect activation of STAT1 by LMP1.
A. B95.8#1 and B95.8#4 BL41 clones were stained with anti IFNR1-PE, IFNRII-PE (filled-in histogram) or isotype controls (open histogram) and analyzed by flow cytometry. B, B95.8#1 and B95.8#4 cells were crosslinked for 0 or 24 hours with/without pre-incubation with either 2 μg/ml of anti-IFNRI and 20 μg/ml of anti-IFN-γ, 22 μg/ml mouse IgG or 1 μM Bay 11-7082. Cells were harvested and lysed in phospholysis buffer and immunoprecipitated for STAT1. Western blotting with anti-p-TyrSTAT1 and anti-STAT1 antibodies was performed. Densitometry is shown below the blot and represents the ratio of p-Tyr-STAT1 to total STAT1. Data is representatitive of three independent experiments.
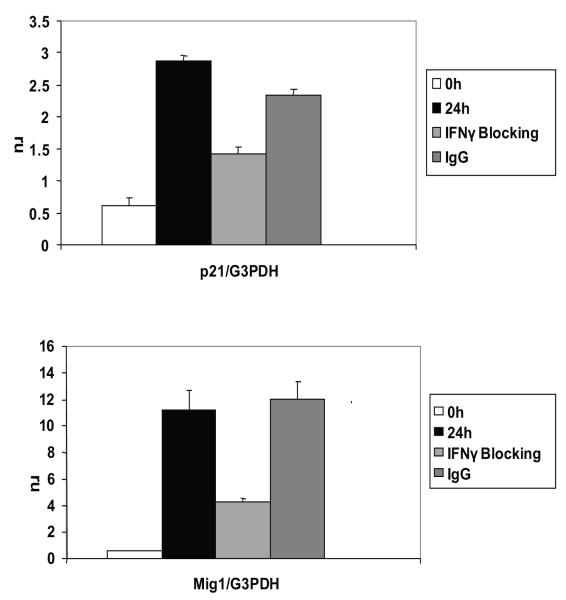
To determine whether blockade of IFN-γ has downstream effects we examined expression of the IFN-γ- and STAT1-inducible genes p21 and monokine induced by interferon-γ (Mig1). Expression of both p21 and Mig1 transcripts was induced by LMP1 after NGFR crosslinking (Fig.7, black bars). Moreover, blockade of the LMP1-induced autocrine IFN-γ pathway significantly decreased expression of p21 (p=0.003) and Mig 1 (p=0.037) (Fig.7, light gray bars). The effect was specific to IFN-γ because addition of control IgG did not significantly affect p21 or Mig1 induction by LMP1 (Fig 7, dark gray bars). Together, these data indicate that LMP1 signaling induces production of IFN-γ that, through an autocrine pathway, induces p-Tyr STAT1 and expression of STAT1-inducible genes.
Figure 7. LMP1-driven STAT1 inducible genes p21 and Mig1 are inhibited by IFNγ blockade.
B95.8#1 cells were crosslinked for 0 (white bars) or 24 hours with (light gray bars) or without (black bars) pre-incubation with anti-IFNR1 and anti-IFN-γ or IgG control (dark gray bars). RNA was isolated with TRIzol and first strand cDNA was generated. Resulting cDNA was used in QPCR with SYBR Green PCR master mix using primers for p21 and Mig1 and GAPDH. Data is shown as the mean relative copy number of p21 or Mig1 compared to GAPDH. Decreases in p21 and Mig1 were statistically significant (p=0.003 and p=0.037, respectively). The results are representative of three independent experiments.
Discussion
We previously reported that the Jak/STAT pathway is constitutively active in PTLD tumors and in EBV+ cell lines derived from PTLD patients (14). However, the role of EBV and the mechanisms driving constitutive Jak/STAT activation remains unclear. Here we show that the EBV protein LMP1 does not directly associate with JAK3 to induce STAT activation in EBV+ B cell lines derived from patients with PTLD. Rather, LMP1 induces production of IFN-γ that, through an autocrine pathway, activates STAT1 downstream of the IFN-γ receptor. Moreover, we show that this pathway is biologically significant, because it is sufficient to elicit expression of STAT1-inducible cellular genes. These studies demonstrate the ability of EBV and LMP1 to influence cellular function through induction of cellular genes.
LMP1 acts as a functional homolog of the CD40 receptor. CD40 can directly interact with, and activate, JAK3 leading to p-Tyr STAT3. The interaction of CD40 with JAK3 occurs via Box1 motifs in CD40, normally required for JAK/receptor interactions. The same JAK-binding motif is also present in the C-terminus of LMP1 raising the possibility that LMP1 may also interact with JAK3 (21). Our analysis of endogenous LMP1 and JAK3 in PTLD-derived EBV+ B lymphoma cell lines, however, revealed that LMP1 and JAK3 do not directly interact although we could readily detect interactions between LMP1 and TRAF3 as well as interactions between JAK3 and γc.
Our study is the first to examine kinetics of STAT activation by LMP1 in B cell lines. Using an inducible LMP1 signaling system we found that activation of STAT1 and STAT3 is delayed and is not a direct effect of LMP1 signaling. Further, the indirect activation of the STAT pathway by LMP1 primarily involves STAT1, as STAT3 activation was either weak or non-existent even after prolonged LMP1 signaling. Therefore, the constitutive STAT3 activation observed in PTLD lines might be regulated differently than STAT1 activation and could potentially involve the autocrine IL-10 axis.
Our data demonstrate that a secreted factor induced by LMP1 is necessary for the indirect activation of STAT1. Indeed we show that LMP1-induced IFN-γ contributes to STAT1 activation because blockade of the IFN-γ receptor and neutralization of IFN-γ with specific antibodies decreased STAT1 p-Tyr to almost background levels. These findings agree with the report of Najjar et al (16) that also argues for indirect activation of STAT1 by IFN-γ in LCL from healthy blood donors. However, those studies also indicated a role for IFN-α in STAT1 activation. In contrast, we found no evidence of IFN-α production by BL41 cells after LMP1 activation. We did detect IL-2 production in supernatants of BL41.NGFR-LMP1 cells after NGFR cross-linking. IL-2 is also known to activate STAT1 and could potentially be contributing to the STAT1 activation we observed. However, blocking antibodies to the IL-2 receptor did affect STAT1 activation in response to LMP1 signaling in BL41.NGFR-LMP1 cells (data not shown). Our studies also differ from Najjar et al. in that they did not directly address the role of LMP1 in activation of STAT1. Instead they utilized EBV+ LCL expressing several other EBV latent genes, in addition to LMP1, that could contribute to the overall STAT activation status.
We also provide the first data on the biologic significance of IFN-γ induction by LMP1 in B cell lymphomas by analyzing expression of STAT1-inducible genes. LMP1 signaling led to Mig1 and p21 gene expression while blockade of IFN-γ significantly decreased the expression levels of both genes. The residual Mig1 and p21 expression may be due to incomplete blockade of the IFN-γ pathway or to other, as yet undefined, pathways of gene activation. With respect to the latter possibility we do not believe IFN-α is involved in STAT1 activation since LMP1 activation did not induce IFN-α production. Although IL-10 is produced by these cells, we do not think it plays a role in STAT1 activation in this model since antibodies to IL-10 do not affect STAT1 activation and since addition of high doses of exogenous IL-10 only weakly activate STAT1. We cannot rule out the possibility that STAT3 activation by IL-10 is contributing to Mig1 and p21 gene expression. Nevertheless, these data underscore the biologic importance of IFN-γ induction and downstream STAT1 activation by LMP1 as they establish, in principal, how this pathway can modulate cellular gene expression. These results also raise the question of the role of Mig1 as and p21 in EBV-infected B cells and PTLD. LMP1 is known to induce several chemokines including IP10, RANTES and MIP1 (22-24). Indeed, we measured production of these chemokines in our analysis of supernatants obtained from BL41.NGFR-LMP1 cells after LMP1 activation (data not shown). Mig1 is an IFN-γ- and STAT1-inducible chemokine that can target activated T cells and induce chemotaxis (25). Additionally, Mig1 can prevent angiogenesis in vivo and is involved in tumor necrosis (26, 27). We show that Mig1 induction in BL41 B lymphoma cells is dependent on LMP1 activation and requires indirect activation of STAT1 through IFN-γ. Based on known functions of Mig1, it is possible that induction of Mig1 by EBV is actually detrimental to lymphoma progression as a consequence of IFN-γ induction by the virus.
p21 is regarded primarily as a cell cycle inhibitor, however in certain circumstances p21 can promote and stabilize CDK4/Cyclin D complex formation leading to G1/S cell cycle progression (28). It has also been suggested that p21 can act as an oncogene and may protect cells from stress induced apoptosis (29, 30). In concordance with the oncogenic role of p21, we have previously shown that SLCL express higher levels of p21 compared to EBV- B cells. Furthermore, we showed that the mTOR inhibitor rapamycin arrests proliferation of SLCL at the G1 stage of the cell cycle and this effect was associated with decreased p21 levels (31). Therefore, it is plausible that in PTLD-associated B cell lymphomas, p21 promotes proliferation and that indirect STAT1 activation may contribute to the overall transforming effects of LMP1. Further studies are necessary to define the role of Mig1 and p21, as well as other STAT1 inducible genes, in PTLD-associated B cell lymphomas. However, our data suggest that targeting STAT1, either through modulating STAT1 levels or inhibiting STAT1 activation by Jak kinases, could potentially provide therapeutic benefit for prevention or treatment of PTLD. In summary, we show that while STAT1 activation by LMP1 is indirect, it can nevertheless participate in downstream gene regulation and therefore may contribute to the pathogenesis of B cell lymphomas and PTLD. Furthermore, understanding the activation of cellular signal transduction pathways by EBV could identify novel therapeutic targets for treatment of PTLD.
Acknowledgments
1Supported by NIH grants RO1 AI41769 (OMM) and CA105157 (OMM). M.V. was partially supported by the Mason Case Fellowship and S.L.L. was supported by an American Cancer Society Fellowship
References
- 1.Kilger E, Kieser A, Baumann M, Hammerschmidt W. Epstein-Barr virus-mediated B-cell proliferation is dependent upon latent membrane protein 1, which simulates an activated CD40 receptor. Embo J. 1998;17(6):1700–1709. doi: 10.1093/emboj/17.6.1700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mainou BA, Everly DN, Jr., Raab-Traub N. Epstein-Barr virus latent membrane protein 1 CTAR1 mediates rodent and human fibroblast transformation through activation of PI3K. Oncogene. 2005;24(46):6917–6924. doi: 10.1038/sj.onc.1208846. [DOI] [PubMed] [Google Scholar]
- 3.Eliopoulos AG, Young LS. Activation of the cJun N-terminal kinase (JNK) pathway by the Epstein-Barr virus-encoded latent membrane protein 1 (LMP1) Oncogene. 1998;16(13):1731–1742. doi: 10.1038/sj.onc.1201694. [DOI] [PubMed] [Google Scholar]
- 4.Dawson CW, Laverick L, Morris MA, Tramoutanis G, Young LS. Epstein-Barr virus-encoded LMP1 regulates epithelial cell motility and invasion via the ERK-MAPK pathway. J Virol. 2008;82(7):3654–3664. doi: 10.1128/JVI.01888-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Eliopoulos AG, Gallagher NJ, Blake SM, Dawson CW, Young LS. Activation of the p38 mitogen-activated protein kinase pathway by Epstein-Barr virus-encoded latent membrane protein 1 coregulates interleukin-6 and interleukin-8 production. J Biol Chem. 1999;274(23):16085–16096. doi: 10.1074/jbc.274.23.16085. [DOI] [PubMed] [Google Scholar]
- 6.Devergne O, McFarland ED Cahir, Mosialos G, Izumi KM, Ware CF, Kieff E. Role of the TRAF binding site and NF-kappaB activation in Epstein-Barr virus latent membrane protein 1-induced cell gene expression. J Virol. 1998;72(10):7900–7908. doi: 10.1128/jvi.72.10.7900-7908.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dawson CW, Tramountanis G, Eliopoulos AG, Young LS. Epstein-Barr virus latent membrane protein 1 (LMP1) activates the phosphatidylinositol 3-kinase/Akt pathway to promote cell survival and induce actin filament remodeling. J Biol Chem. 2003;278(6):3694–3704. doi: 10.1074/jbc.M209840200. [DOI] [PubMed] [Google Scholar]
- 8.Gires O, Kohlhuber F, Kilger E, Baumann M, Kieser A, Kaiser C, et al. Latent membrane protein 1 of Epstein-Barr virus interacts with JAK3 and activates STAT proteins. EMBO J. 1999;18(11):3064–3073. doi: 10.1093/emboj/18.11.3064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Higuchi M, Kieff E, Izumi KM. The Epstein-Barr virus latent membrane protein 1 putative Janus kinase 3 (JAK3) binding domain does not mediate Jak3 association or activation in B lymphoma or lymphoblastoid cell lines. Journal of Virology. 2002;76(1):455–459. doi: 10.1128/JVI.76.1.455-459.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gupta S, Yan H, Wong LH, Ralph S, Krolewski J, Schindler C. The SH2 domains of Stat1 and Stat2 mediate multiple interactions in the transduction of IFN-alpha signals. Embo J. 1996;15(5):1075–1084. [PMC free article] [PubMed] [Google Scholar]
- 11.Remy I, Wilson IA, Michnick SW. Erythropoietin receptor activation by a ligand-induced conformation change. Science. 1999;283(5404):990–993. doi: 10.1126/science.283.5404.990. [DOI] [PubMed] [Google Scholar]
- 12.Barahmand-Pour F, Meinke A, Groner B, Decker T. Jak2-Stat5 interactions analyzed in yeast. J Biol Chem. 1998;273(20):12567–12575. doi: 10.1074/jbc.273.20.12567. [DOI] [PubMed] [Google Scholar]
- 13.Shuai K, Horvath CM, Huang LH, Qureshi SA, Cowburn D, Darnell JE., Jr. Interferon activation of the transcription factor Stat91 involves dimerization through SH2-phosphotyrosyl peptide interactions. Cell. 1994;76(5):821–828. doi: 10.1016/0092-8674(94)90357-3. [DOI] [PubMed] [Google Scholar]
- 14.Nepomuceno RR, Snow AL, Beatty PR, Krams SM, Martinez OM. Constitutive activation of Jak/STAT proteins in Epstein-Barr virus B cell lines from patients with posttransplant lymphoproliferative disorder. Transplantation. 2002;74(3):396–402. doi: 10.1097/00007890-200208150-00017. [DOI] [PubMed] [Google Scholar]
- 15.Nepomuceno RR, Balatoni CE, Natkunam Y, Snow AL, Krams SM, Martinez OM. Rapamycin inhibits the interleukin 10 signal transduction pathway and the growth of Epstein Barr virus B-cell lymphomas. Cancer Res. 2003;63(15):4472–4480. [PubMed] [Google Scholar]
- 16.Najjar I, Baran-Marszak F, Le Clorennec C, Laguillier C, Schischmanoff O, Youlyouz-Marfak I, et al. Latent membrane protein 1 regulates STAT1 through NF-kappaB-dependent interferon secretion in Epstein-Barr virus-immortalized B cells. J Virol. 2005;79(8):4936–4943. doi: 10.1128/JVI.79.8.4936-4943.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lambert SL, Martinez OM. Latent membrane protein 1 of EBV activates phosphatidylinositol 3-kinase to induce production of IL 10. J Immunol. 2007;179(12):8225–8234. doi: 10.4049/jimmunol.179.12.8225. [DOI] [PubMed] [Google Scholar]
- 18.Sandvej K, Gratama JW, Munch M, Zhou XG, Bolhuis RL, Andresen BS, et al. Sequence analysis of the Epstein-Barr virus (EBV) latent membrane protein-1 gene and promoter region: identification of four variants among wild-type EBV isolates. Blood. 1997;90(1):323–330. [PubMed] [Google Scholar]
- 19.Beatty PR, Krams SM, Martinez OM. Involvement of IL-10 in the autonomous growth of EBV-transformed B cell lines. J Immunol. 1997;158(9):4045–4051. [PubMed] [Google Scholar]
- 20.Mavropoulos A, Sully G, Cope AP, Clark AR. Stabilization of IFN-gamma mRNA by MAPK p38 in IL-12- and IL-18-stimulated human NK cells. Blood. 2005;105(1):282–288. doi: 10.1182/blood-2004-07-2782. [DOI] [PubMed] [Google Scholar]
- 21.Hanissian SH, Geha RS. Jak3 is associated with CD40 and is critical for CD40 induction of gene expression in B cells. Immunity. 1997;6:379–387. doi: 10.1016/s1074-7613(00)80281-2. [DOI] [PubMed] [Google Scholar]
- 22.Buettner M, Meyer B, Schreck S, Niedobitek G. Expression of RANTES and MCP-1 in epithelial cells is regulated via LMP1 and CD40. Int J Cancer. 2007;121(12):2703–2710. doi: 10.1002/ijc.23018. [DOI] [PubMed] [Google Scholar]
- 23.Li J, Zhang XS, Xie D, Deng HX, Gao YF, Chen QY, et al. Expression of immune-related molecules in primary EBV-positive Chinese nasopharyngeal carcinoma: associated with latent membrane protein 1 (LMP1) expression. Cancer Biol Ther. 2007;6(12):1997–2004. doi: 10.4161/cbt.6.12.5160. [DOI] [PubMed] [Google Scholar]
- 24.Nakayama T, Hieshima K, Nagakubo D, Sato E, Nakayama M, Kawa K, et al. Selective induction of Th2-attracting chemokines CCL17 and CCL22 in human B cells by latent membrane protein 1 of Epstein-Barr virus. J Virol. 2004;78(4):1665–1674. doi: 10.1128/JVI.78.4.1665-1674.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liao F, Rabin RL, Yannelli JR, Koniaris LG, Vanguri P, Farber JM. Human Mig chemokine: biochemical and functional characterization. J Exp Med. 1995;182(5):1301–1314. doi: 10.1084/jem.182.5.1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Teruya-Feldstein J, Jaffe ES, Burd PR, Kanegane H, Kingma DW, Wilson WH, et al. The role of Mig, the monokine induced by interferon-gamma, and IP-10, the interferon-gamma-inducible protein-10, in tissue necrosis and vascular damage associated with Epstein-Barr virus- positive lymphoproliferative disease. Blood. 1997;90(10):4099–4105. [PubMed] [Google Scholar]
- 27.Strieter RM, Polverini PJ, Arenberg DA, Kunkel SL. The role of CXC chemokines as regulators of angiogenesis. Shock. 1995;4(3):155–160. doi: 10.1097/00024382-199509000-00001. [DOI] [PubMed] [Google Scholar]
- 28.LaBaer J, Garrett MD, Stevenson LF, Slingerland JM, Sandhu C, Chou HS, et al. New functional activities for the p21 family of CDK inhibitors. Genes Dev. 1997;11(7):847–862. doi: 10.1101/gad.11.7.847. [DOI] [PubMed] [Google Scholar]
- 29.De la Cueva E, Garcia-Cao I, Herranz M, Lopez P, Garcia-Palencia P, Flores JM, et al. Tumorigenic activity of p21Waf1/Cip1 in thymic lymphoma. Oncogene. 2006;25(29):4128–4132. doi: 10.1038/sj.onc.1209432. [DOI] [PubMed] [Google Scholar]
- 30.Gartel AL, Tyner AL. The role of the cyclin-dependent kinase inhibitor p21 in apoptosis. Mol Cancer Ther. 2002;1(8):639–649. [PubMed] [Google Scholar]
- 31.Vaysberg M, Balatoni CE, Nepomuceno RR, Krams SM, Martinez OM. Rapamycin inhibits proliferation of Epstein-Barr virus-positive B-cell lymphomas through modulation of cell-cycle protein expression. Transplantation. 2007;83(8):1114–1121. doi: 10.1097/01.tp.0000260142.38619.9c. [DOI] [PubMed] [Google Scholar]