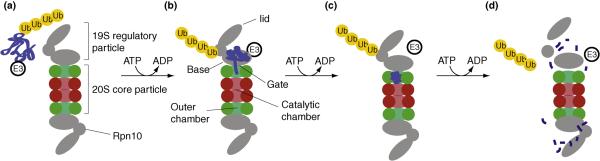
Figure 4. A model of the substrate degradation by the proteasome.
The degradation process is part of a larger pathway, consisting of activation of ubiquitin, conjugation of the protein substrate and ubiquitins, degradation of the tagged protein, and deubiquitination to recycle the ubiquitins (bottom). The degradation process (top) is modeled by four key states and transitions between them, discussed in more detail in the text. The modeled system involves the 26S proteasome, the E3 ligase enzyme and the ubiquinated substrate protein. The four key states of the model are: [A] stable holo 26S, [B] recruitment of polyubiquinated substrate, [C] storage of substrate inside the proteasome and [D] disassociated and disassembled regulator particle. Arrows show transitions between states. As more information is obtained, the model can become more detailed by adding key states, increasing the spatial resolution of each key state, and mapping trajectories between key states.