Figure 1.
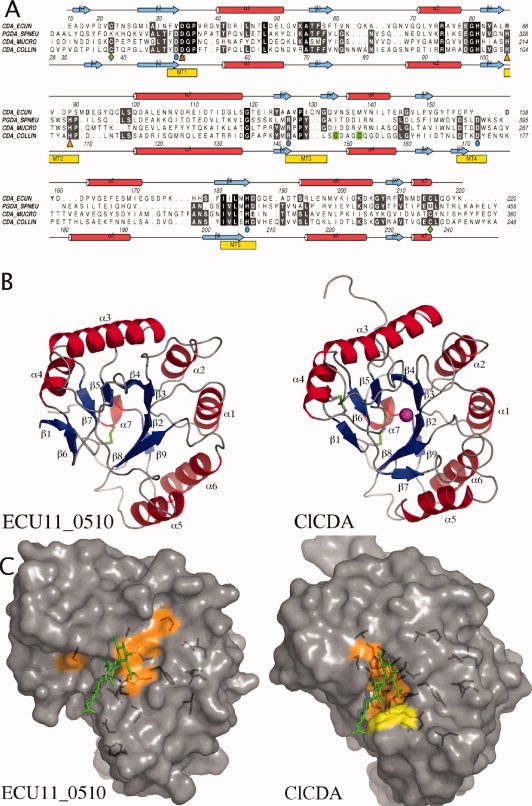
(A) A structure-based sequence alignment. Secondary structure of the ClCDA and the ECU11_0510 proteins are shown; α helixes are colored red and β strands in blue. Metal-coordinating residues are highlighted with orange triangles. Catalytic residues are highlighted with cyan circles. Conserved disulphide bridges are highlighted with green diamonds. A second nonconserved disulphide bridge in ClCDA is highlighted in green. CDA-ECCUN = ECU11_0510, PGDA_SPNEU = Streptococcus pneumoniae PgdA, CDA_MUCRO = Mucor Rouxii Chitin Deacetylase and CDA_COLLIN = Colletotrichum lindemuthianum Chitin Deacetylase. (B) Overall structure comparison of Encephalitozoon cuniculi putative chitin deacetylase, ECU11_0510 (left panel), and ClCDA (right panel). α helixes are colored red and β strands in blue. Conserved disulphide bridges are highlighted in green. Zinc in the active site of the ClCDA is shown as a magenta sphere. (C) ECU11_0510 and ClCDA are shown in a surface representation. A chitotrioside carrying the previously proposed oxyanion reaction intermediate was docked into the ClCDA active site as described previously21 and then superposed onto the active sites of the ECU11_0510. The consensus sequence was generated by aligning 12 CE4 esterase sequences using T-coffee. Residues that are identical or show conservative substitutions and are present in 11/12 sequences are represented on the surface in orange and yellow, respectively.
