Abstract
About 85% of the maize genome consists of highly repetitive sequences that are interspersed by low-copy, gene-coding sequences. The maize community has dealt with this genomic complexity by the construction of an integrated genetic and physical map (iMap), but this resource alone was not sufficient for ensuring the quality of the current sequence build. For this purpose, we constructed a genome-wide, high-resolution optical map of the maize inbred line B73 genome containing >91,000 restriction sites (averaging 1 site/∼23 kb) accrued from mapping genomic DNA molecules. Our optical map comprises 66 contigs, averaging 31.88 Mb in size and spanning 91.5% (2,103.93 Mb/∼2,300 Mb) of the maize genome. A new algorithm was created that considered both optical map and unfinished BAC sequence data for placing 60/66 (2,032.42 Mb) optical map contigs onto the maize iMap. The alignment of optical maps against numerous data sources yielded comprehensive results that proved revealing and productive. For example, gaps were uncovered and characterized within the iMap, the FPC (fingerprinted contigs) map, and the chromosome-wide pseudomolecules. Such alignments also suggested amended placements of FPC contigs on the maize genetic map and proactively guided the assembly of chromosome-wide pseudomolecules, especially within complex genomic regions. Lastly, we think that the full integration of B73 optical maps with the maize iMap would greatly facilitate maize sequence finishing efforts that would make it a valuable reference for comparative studies among cereals, or other maize inbred lines and cultivars.
Author Summary
The maize genome contains abundant repeats interspersed by low-copy, gene-coding sequences that make it a challenge to sequence; consequently, current BAC sequence assemblies average 11 contigs per clone. The iMap deals with such complexity by the judicious integration of IBM genetic and B73 physical maps, but the B73 genome structure could differ from the IBM population because of genetic recombination and subsequent rearrangements. Accordingly, we report a genome-wide, high-resolution optical map of maize B73 genome that was constructed from the direct analysis of genomic DNA molecules without using genetic markers. The integration of optical and iMap resources with comparisons to FPC maps enabled a uniquely comprehensive and scalable assessment of a given BAC's sequence assembly, its placement within a FPC contig, and the location of this FPC contig within a chromosome-wide pseudomolecule. As such, the overall utility of the maize optical map for the validation of sequence assemblies has been significant and demonstrates the inherent advantages of single molecule platforms. Construction of the maize optical map represents the first physical map of a eukaryotic genome larger than 400 Mb that was created de novo from individual genomic DNA molecules.
Introduction
Maize (Zea mays ssp. mays L.) is a pervasive, economically valuable crop supplying the world with food, animal feed, and with biofeedstocks used in the synthesis of a broad range of industrial products. It is also a model system for classical genetics and cytogenetics that has significantly contributed to our understanding of fundamental processes that include reproduction, photosynthesis, biosynthesis of primary metabolites, mobile elements, and chromosome structure-function relationships. Investigators have developed extensive genetic tools over the last two decades dealing with male sterility, QTLs, regeneration of crop species, wide hybridization, marker assisted selection, associative mapping, endosperm mutants, transgenic crops, genetic control of meiosis, transposable elements, chromosome elimination, etc. In addition, diverse germplasms have been accumulated that have leveraged the assessments of genomic modifications during domestication, molecular mechanisms of heterosis, and the roles played by mobile DNA elements affecting genome evolution. Such advances are now being rapidly exploited with paradigm shifting tools and resources that are fostering insights emerging from fully sequenced and annotated genomes. In 2005 three funding agencies – NSF, DOE and USDA – jointly pledged $32 million towards a 4-year program to sequence the maize genome. These agencies' goals were to ensure that cutting-edge genomic resources would be available for maize to accelerate translational research in the agriculture and bioenergy sectors.
The maize genome is estimated to be 2.3–2.5 gigabases (Gb) in size [1], and its architecture presents significant challenges for comprehensive sequencing. An intriguing attribute of the maize genome is its allotetraploidy nature that originated at least 5 million years ago (mya) from two progenitors, which had previously diverged from a common ancestor about 12 mya [2]–[3]. The maize genome underwent a whole genome duplication event in the hybridization of the two progenitors, and then gradually became diploid through loss of ∼50% of one of its progenitors' gene copies [4]–[8].
The architecture of the maize genome is also heavily punctuated by a complex motif of repetitive elements. About 85% of the genome is made up of a complex mix of repetitive DNA that mainly includes numerous families of retrotransposons such as Tekay, Huck, PREM-2, Opie, Ji, etc. [9]–[11]. These retroelements mostly appeared during the last 1 to 3 million years and thus show great similarity [9]. The chromosome “knobs” consist of megabase-sized satellite sequences interspersed with retrotransposons, while the euchromatic regions harbor repetitive insertions of transposons, with most retrotransposons tending to insert within each other, resulting in nested retrotransposons in the intergenic regions [9], [11]–[17]. Therefore, maize genes are like small islands surrounded by seas of nested retrotransposons, and such challenging attributes have necessitated development of multiple sequencing approaches.
Given the current need for a broadly informative representation that includes coding sequences and precise physical characterization of gaps between genes, accurate genetic and physical maps are required for guiding the large-scale sequencing of maize genome. The genetic, physical, and integrated maps available for maize are briefly described. The 1935 maize genetic map featured just 62 loci that relied on morphological variants [18]. Advancements in new technologies and genomic insights led to the addition of nearly 6,000 markers to create a high resolution genetic map using the intermated B73 X Mo17 (IBM) populations [19]–[20] (http://maize-mapping.plantgenomics.iastate.edu/). This augmentation drew new resources from the development of cytological markers based on B-A translocations, molecular markers based on isozymes, restriction fragment length polymorphisms (RFLPs), microsatellite or simple sequence repeat (SSR) markers, single nucleotide polymorphisms (SNPs), and cDNA or expressed sequence tags (EST) markers [19], [21]–[38]. In addition, FPC (fingerprinted contigs) [39] map contigs were also anchored to this genetic linkage map, and this highly integrated resource became known as the “iMap.”
Early physical mapping of maize used a YAC (yeast artificial chromosome) library constructed from an inbred line UE95 [34]. The YAC libraries proved to be of limited utility due to a significant level of clone chimerism, or issues surrounding YAC stability and faithful representation of genome copy number [40]. With the advent of stable large-insert cloning in bacteria, more reliable BAC (bacterial artificial chromosome) [41] libraries were constructed for the maize B73 inbred line. Clones were fingerprinted, hybridized with known molecular markers (genetic), and FPC mapped. These efforts integrated the genetic and physical maps through assignment of molecular markers from the genetic map to individual BAC clones within FPC contigs [35], [42]–[46]. FPC maps were later greatly refined by HICF (fluorescent-based high-information-content fingerprinting) mapping of these libraries which reduced the number of FPC contigs from 4,518 to 1,500 [47]. The number of FPC contigs was further reduced to 721 (2,150 Mb; May, 2006) by manual curation based on agarose-based fingerprinting and on knowledge gleaned from the HICF map and syntenic markers between the maize and rice genomes [37]. In addition to the ∼6,000 genetic markers, there are over 24,000 sequence markers integrated into the maize genetic-physical (FPC) map (also termed iMap), including expressed sequence tag (EST)-derived unigene markers, overgos derived from maize EST sequences, conserved genomic sequences, and end-sequence data from gene-containing BACs. The inclusion of these sequence markers into the integrated map (iMap) (IBM2; iMap; http://www.maizemap.org/iMapDB/iMap.html) has greatly increased the marker density across the entire maize genome and created a framework for directed clone-based sequencing and assembly of chromosome-wide pseudomolecules [37],[44],[48]. However, the maize genome is structurally highly polymorphic, as seen in the significant structural variation among different inbred lines and even between different haplotypes [49]–[52]. Because the maize iMap integrates the IBM genetic map with the B73 inbred line FPC physical map, structural differences between the IBM population and the B73 genome (targeted genome for sequencing http://ftp.maizesequence.org/release-3b.50/All Releases/ [53]) would be expected. The primary construction of high-resolution physical maps that are not dependent on the IBM genetic map would offer an essential resource for the comprehensive and accurate assembly of the maize B73 reference genome.
Sequencing efforts for the maize genome have progressed through three stages: the pilot, gene enrichment, and clone-by-clone full genome sequencing stages. 1) The pilot sequencing effort considered large parts of chromosome arms and BAC-end sequence gathered from random clones; this provided an early glimpse into genome structure, organization, and sequence composition [48], [54]–[55]. 2) The gene-enrichment approaches culled gene-rich templates for side-stepping notoriously complex sequence repeats and high copy number DNA elements present in the maize genome. Enrichment was accomplished by a variety of sequencing approaches that included ESTs, genome filtration (methylation filtration and high-Cot selection), RescueMu (RM), and hypomethylated partial restriction (HMPR) [56]–[63]. Sequence data enriched for genes, collectively termed as Genome Survey Sequences (GSSs), are scattered throughout the maize genome, typically comprising small sequence contigs a few kilobases in size [61]. 3) In contrast, clone-by-clone sequencing used a comprehensive, hierarchical, map-based approach that allowed construction of a BAC minimal tiling path across the iMap. Tiled BACs were then individually shotgun-sequenced and assembled [12], [64]–[66]. Although this map-based approach simplified assembly, an individual BAC assembly typically contained multiple unordered sequence contigs. Sequencing of the maize genome is now in the finishing phase with more than 16,000 sequenced BACs [53]. But complete sequencing and creation of a highly accurate assembly of the maize genome still hold daunting challenges for the maize community.
A direct and encompassing way to deal with the formidable architecture of the maize genome is to analyze “chunks” of it, at high-resolution, that are as large as possible. In this way, nests of sequence repeats are largely bridged by chunks that offer a sufficient level of unique sequence information for supporting de novo genome assembly. With this concept in mind, we constructed a high-resolution optical map [67]–[76] that spans ∼91% of the maize genome by the de novo assembly of a large data set containing ordered restriction maps of individual genomic DNA molecules ∼500 kb in size. This ordered restriction map provides an independent resource that lays out an accurate physical metric across the entire maize genome. Because large molecules were analyzed, we were able to physically map repeat-rich regions and link sequence and map data within complex genomic regions. We show here that our maize optical map identifies gaps within and between sequence contigs and guides the assembly and validation of reference chromosomes.
Results
Data acquisition and map assembly
We constructed a whole-genome shotgun optical map for maize using the CpG methylation insensitive restriction enzyme SwaI. The optical map data set contains 2,116,074 genomic DNA molecules, ranging in size from 300 kb to 3,700 kb, and totaling ∼927,604 Mb, or ∼403× coverage of the maize genome. The maps in this raw data set—one optical restriction map per genomic DNA molecule—have a mean length of 438.4 kb with an average fragment size of 26.1 kb.
Because of the vast size of the maize optical data set, our de novo assembly of maps relied on a divide and conquer strategy that leveraged available cluster computing resources [77]. Briefly, we divided the raw map data set into 40 separate bins. Each bin was assembled into contigs and processed to remove redundant contigs and/or overlapping contigs, producing seed maps (consensus maps) for our iterative assembly scheme (Materials and Methods). After five initial cycles of iterative assembly, the terminal 40 restriction fragments of a seed map (Materials and Methods) were selected for augmentation of optical contigs that were >10 Mb. These optical consensus maps were lengthened and their depth of coverage was increased through an additional 15 cycles of iterative assembly using the entire map data set. In this way, we constructed 66 optical consensus maps spanning a total of 2,103.93 Mb.
The consensus maps were internally validated in an additional iterative assembly step. They were partitioned into a series of overlapping 1 Mb map intervals for use as new seed maps, with the overlaps covering ∼500 kb. Because this diagnostic assembly reproduced the original set of 66 parental contigs, the current optical assembly is apparently free of any chimeric maps. Statistics describing the 66 optical map contigs are shown in Table 1. In total, 339,280 of the 2,116,074 maps were assembled into 66 optical map contigs. The average depth of coverage is 72 restriction fragments per contig (Table 1). The breadth of these contigs range from 3.64 Mb–100.76 Mb, and the average contig size is 31.88 Mb. The average size of restriction fragments of each contig ranges from 21.32 kb to 28.53 kb, with the overall size averaging 23.56 kb (Table 1). Lastly, the rate of contig formation was 16.03%, and we attribute this modest value to the modest rate of restriction digestion caused by unknown inhibitors within our DNA preps (genomic; ∼500 kb sized molecules) that attenuated restriction enzyme action. We leveraged the assembly process for overcoming this problem by increasing the number of digested molecules within the raw data set for biasing those molecules with adequate restriction patterns supporting confident contig formation.
Table 1. Statistics of optical map contigs and the anchoring of FPC contigs.
| Optical Map Contig Name | Contig Size (Mb) | Ave Frag Size (kb) | # of Sin Mol Maps | Coverage (X) | Ave. SD | Chr. Anchored | FPC Contigs Spanned |
| OMcontig_0 | 100.76 | 22.82 | 15532 | 71.75 | 2.37 | 1 | ctg30–48 |
| OMcontig_1 | 97.04 | 22.17 | 15903 | 76.15 | 2.34 | 4 | ctg176–196 |
| OMcontig_2 | 94.49 | 23.30 | 15048 | 75.14 | 2.38 | 2 | ctg68–84 |
| OMcontig_3 | 85.78 | 22.45 | 14382 | 77.58 | 2.37 | 1 | ctg12–30 |
| OMcontig_4 | 84.69 | 22.83 | 13331 | 73.39 | 2.34 | 8 | ctg332–354 |
| OMcontig_5 | 83.37 | 22.42 | 13010 | 72.46 | 2.32 | 2 | ctg87–98 |
| OMcontig_6 | 70.90 | 22.85 | 12456 | 81.35 | 2.40 | 6 | ctg279–291 |
| OMcontig_7 | 68.92 | 22.55 | 12094 | 80.65 | 2.39 | 5 | ctg204–222 |
| OMcontig_8 | 65.88 | 22.28 | 11397 | 80.45 | 2.33 | 10 | ctg405–420 |
| OMcontig_9 | 58.50 | 23.34 | 8773 | 70.35 | 2.42 | 4 | ctg169–176 |
| OMcontig_10 | 57.33 | 24.52 | 9081 | 73.21 | 2.36 | 7 | ctg301–315 |
| OMcontig_11 | 56.78 | 23.74 | 9156 | 75.18 | 2.43 | 3 | ctg132–149 |
| OMcontig_12 | 55.30 | 23.88 | 10278 | 86.38 | 2.44 | 3 | ctg121–131 |
| OMcontig_13 | 55.02 | 22.85 | 9120 | 77.25 | 2.39 | 3 | ctg111–118 |
| OMcontig_14 | 52.11 | 23.46 | 8026 | 71.65 | 2.43 | 5 | ctg232–238 |
| OMcontig_15 | 50.50 | 22.27 | 8426 | 78.10 | 2.35 | 1 | ctg1–12 |
| OMcontig_16 | 49.65 | 23.32 | 8204 | 77.72 | 2.43 | 8 | ctg326–334 |
| OMcontig_17 | 49.87 | 23.82 | 7795 | 73.13 | 2.49 | 4 | ctg196–197/ctg376–381 |
| OMcontig_18 | 46.99 | 23.05 | 7207 | 71.59 | 2.35 | 9 | ctg371–376 |
| OMcontig_19 | 47.25 | 22.50 | 8293 | 81.33 | 2.36 | 1 | ctg51–64 |
| OMcontig_20 | 43.23 | 23.17 | 6762 | 72.89 | 2.43 | 7 | ctg292–300 |
| OMcontig_21 | 40.29 | 22.25 | 7508 | 86.06 | 2.36 | 10 | ctg392–398 |
| OMcontig_22 | 38.60 | 23.87 | 5760 | 69.53 | 2.44 | 5 | ctg223–231 |
| OMcontig_23 | 38.23 | 23.18 | 6946 | 84.95 | 2.39 | 9 | ctg383–391 |
| OMcontig_24 | 33.63 | 22.48 | 5434 | 75.07 | 2.38 | 4 | ctg160–164 |
| OMcontig_25 | 29.78 | 22.80 | 4650 | 72.85 | 2.32 | 5 | ctg238–247 |
| OMcontig_26 | 27.71 | 23.76 | 4893 | 82.23 | 2.39 | 2 | ctg99–104 |
| OMcontig_27 | 26.19 | 22.83 | 4524 | 80.34 | 2.33 | 7 | ctg317–321 |
| OMcontig_28 | 25.16 | 23.70 | 3491 | 64.90 | 2.42 | 3 | ctg121/255 |
| OMcontig_29 | 24.54 | 23.53 | 3940 | 74.08 | 2.39 | 4 | ctg170–171 |
| OMcontig_30 | 24.16 | 21.59 | 4135 | 78.77 | 2.19 | 6 | ctg267–269 |
| OMcontig_31 | 23.93 | 21.69 | 4254 | 82.07 | 2.39 | 7 | ctg322–325 |
| OMcontig_32 | 23.15 | 22.88 | 4041 | 81.14 | 2.35 | 8 | ctg354–358 |
| OMcontig_33 | 22.99 | 22.08 | 3921 | 79.63 | 2.33 | 5 | ctg248–254 |
| OMcontig_34 | 21.07 | 23.46 | 3421 | 75.37 | 2.36 | ||
| OMcontig_35 | 20.54 | 23.89 | 3269 | 75.73 | 2.54 | 8 | ctg359–366 |
| OMcontig_36 | 20.51 | 22.48 | 3457 | 78.10 | 2.27 | 6 | ctg265–269 |
| OMcontig_37 | 20.32 | 23.52 | 2882 | 66.75 | 2.40 | 10 | ctg399–401 |
| OMcontig_38 | 16.98 | 22.37 | 2766 | 76.90 | 2.37 | 4 | ctg199–203 |
| OMcontig_39 | 16.13 | 21.85 | 3174 | 90.19 | 2.34 | 2 | ctg108–110 |
| OMcontig_40 | 15.61 | 24.98 | 2272 | 67.09 | 2.40 | unknown | ctg430 |
| OMcontig_41 | 14.53 | 24.17 | 1831 | 58.63 | 2.48 | unknown | ctg449 |
| OMcontig_42 | 14.31 | 24.02 | 1981 | 64.32 | 2.48 | 5 | ctg231–232 |
| OMcontig_43 | 13.48 | 23.48 | 1775 | 61.34 | 2.30 | 7 | ctg300–301 |
| OMcontig_44 | 13.34 | 22.01 | 1989 | 70.50 | 2.36 | 4 | ctg156–159 |
| OMcontig_45 | 13.45 | 22.91 | 2001 | 70.16 | 2.35 | 10 | ctg401–404 |
| OMcontig_46 | 13.29 | 22.99 | 2069 | 72.28 | 2.38 | 3 | ctg118–120 |
| OMcontig_47 | 12.73 | 21.32 | 2088 | 75.57 | 2.22 | 3 | ctg150 |
| OMcontig_48 | 11.86 | 27.91 | 1448 | 58.58 | 2.60 | 6 | ctg256–260 |
| OMcontig_49 | 11.49 | 25.64 | 1478 | 60.39 | 2.51 | 2 | ctg84 |
| OMcontig_50 | 10.34 | 25.22 | 1266 | 57.16 | 2.59 | 2 | ctg120 |
| OMcontig_51 | 10.33 | 23.05 | 1475 | 65.42 | 2.38 | 9 | ctg368–370 |
| OMcontig_53 | 8.70 | 24.51 | 1255 | 66.65 | 2.43 | 6 | ctg271–274 |
| OMcontig_55 | 8.82 | 25.28 | 1043 | 55.96 | 2.48 | unknown | ctg448 |
| OMcontig_56 | 7.89 | 26.03 | 965 | 58.37 | 2.43 | 1 | ctg49 |
| OMcontig_57 | 7.84 | 23.19 | 1103 | 65.17 | 2.39 | ||
| OMcontig_58 | 6.17 | 26.02 | 683 | 51.11 | 2.51 | 2 | ctg106–108 |
| OMcontig_59 | 5.68 | 22.81 | 939 | 77.11 | 2.40 | 1 | ctg50 |
| OMcontig_60 | 5.72 | 21.60 | 885 | 70.21 | 2.37 | 6 | ctg267 |
| OMcontig_61 | 5.69 | 23.81 | 911 | 76.34 | 2.45 | 1 | ctg65–67 |
| OMcontig_64 | 4.52 | 27.08 | 491 | 51.02 | 2.72 | 2 | ctg106 |
| OMcontig_65 | 4.16 | 24.47 | 521 | 58.40 | 2.45 | 6 | ctg264 |
| OMcontig_66 | 4.14 | 28.53 | 593 | 66.00 | 2.72 | 2 | ctg105 |
| OMcontig_67 | 3.89 | 22.90 | 565 | 64.92 | 2.30 | 6 | ctg263 |
| OMcontig_68 | 4.04 | 27.11 | 487 | 56.27 | 2.59 | 7 | ctg321–322 |
| OMcontig_69 | 3.64 | 26.37 | 426 | 54.63 | 2.62 | ||
| Total/ Ave . | 2103.93/ 31.88 | 23.56 | 339280 | 71.61 | 2.41 |
*Note: Ave Frag Size = average fragment size, Sin Mol Map = single molecule map, Ave. SD = average standard deviation, Chr. = chromosome.
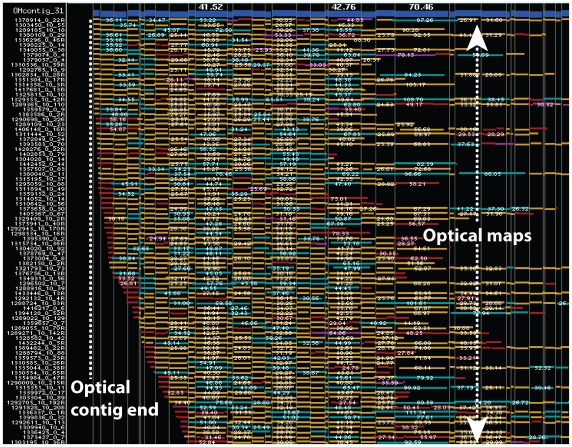
Optical contigs terminate to form a gap when the SwaI restriction site density is low, or when a contig reaches the end of a chromosome. Sharply demarcated contig edges may represent telomere associated sequences near chromosome ends. Using these criteria we identified 15 contigs (OMcontigs_7, 8, 13, 16, 20, 21, 23, 28, 31, 35, 38, 39, 47, 51, and 61) that have reached the ends of chromosome as evidenced by contig “edges” comprising more than 5 maps that show no significant map “overhangs” (Figure 1). A collection of DNA molecules (maps) is said to overhang at a contig's end when their terminal restriction fragments are large and vary in length—such patterns describe gaps.
Figure 1. A screenshot of an optical contig (OMcontig_31) showing a possible telomeric-end.
The Genspect viewer depicts each optical map, constructed from a genomic DNA molecule, as a horizontal track consisting of colored boxes. The length of each box represents the size (in kb) of a restriction fragment within a genomic molecule. The map information of the entire contig is combined into a single track (top; blue) called the consensus map. Restriction fragments are colored keyed for indicating their agreement with the consensus map; gold boxes show agreement, while red (false cut), blue (missing cut), and purple (false cut) indicate aforementioned restriction map differences. This deep contig shows a distinct edge, or end, populated by ∼40 optical maps indicating a telomeric region (Chr7).
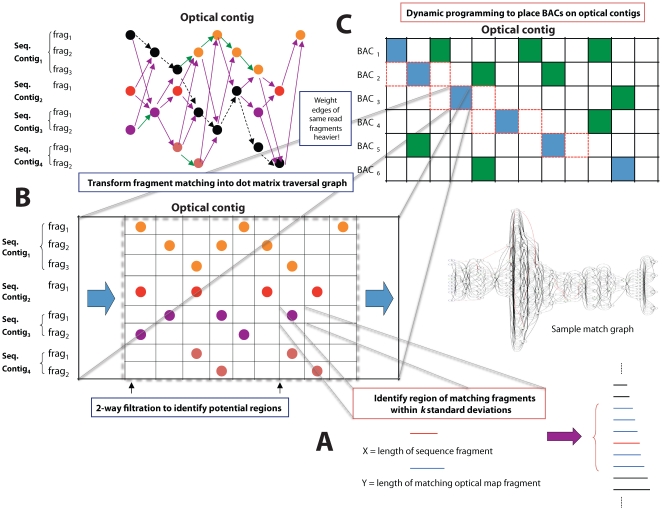
Development of a new algorithm, BACop, for integration of optical map contigs with iMap
The maize genome optical map contains 66 optical contigs and 91,453 ordered SwaI restriction fragments. However, placement of tiled (16,848 FPC clones), but unfinished, BAC sequences released by the maize genome sequencing project (release 3b.50; http://ftp.maizesequence.org/release-3b.50/All%20Releases/; March 19, 2009) on optical maps required development of a new algorithm that considers alignments of FPC clones comprising unordered and unoriented sequence contigs (averaging 11 sequence contigs per BAC). We had developed a new algorithm several years ago to integrate the optical and FPC maps through the alignment of unfinished BAC sequence data (Materials and Methods, Figure 2). Our motivation for its development was to anchor large optical contigs to the iMap, which in 2007 contained only ∼6,000 sequenced BACs. The algorithm—named “BACop” —considers “complete” SwaI restriction fragments (fragments having pairs of SwaI sites) present in the in silico digest of BAC sequence data; the BACs that are analyzed are restricted to those placed on the FPC map. When several consecutive restriction fragments are present, BACop places a set of contigs, belonging to a BAC, onto the optical contig using boundaries consistent with the upper size range (250 kb) of such clones. This alignment also considers the fragment sizing error model used for alignment of optical and sequence in silico maps [78]. The final placement of optical map contigs onto the maize iMap relies on global considerations of BAC locations on the optical vs. FPC maps (Figure 2). For example, when both maps have placed BACs showing similar ordering and spacing (with 20% error allowed), alignments are said to be “co-linear.” Overlaps or gaps are represented on the FPC framework when discordant optical and FPC distances range from ∼200 kb to 2 Mb. When an optical contig aligns to multiple locations on the iMap, the alignment having the greatest number of BACs is selected.
Figure 2. BACop, an algorithm that anchors optical contigs onto the iMap.
BACop (Materials and Methods) employs four distinct steps for anchoring optical contigs; we illustrate the first three steps here: (A) Restriction fragments of an optical contig map are matched against BAC sequences comprising multiple sub-contigs. (B) Matching BAC sequence contigs are located along the optical contig map. (C) Dynamic programming places BACs onto optical map contigs. Seq. = sequence, frag = restriction fragment, and BAC = bacterial artificial chromosome.
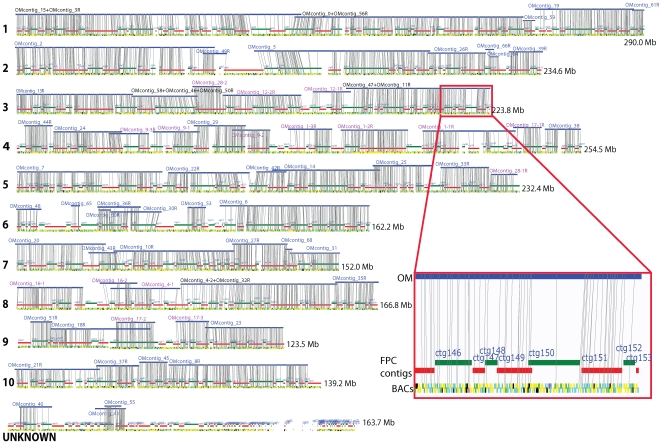
BACop placed 91% of the optical contigs (60/66) onto the 2006 FPC map [37], with 3 additional optical contigs placed onto FPC contigs that lack chromosome assignments (Table 1; Figure 3). The total breadth (2,032.42 Mb) of 60 optical contigs placed on this FPC map (1,981 Mb) is slightly larger than its total size. This extra mass accrues from optical contigs that bridge across FPC gaps, and pairs of optical contigs that partially span gaps. At these locations FPC gaps (reported, or optically revealed) are apparent because one of the overlapping optical contigs in such pairs has very few if any placed BACs, indicating the presence of a large gap between adjoining FPC contigs, or their incorrect placement. For example, optical contigs OMcontigs_28 and 50 were originally incorrectly placed onto Chr 3 FPC contigs ctg120 and ctg121. Although these optical contigs overlapped, only OMcontig_28 showed a dense pattern of BACs that aligned to FPC contig ctg121, but none to the adjoining FPC ctg120 within the overlap region. After realigning each half of OMcontig_28, the half that hadn't aligned was found to align to the end of the chromosome 5 FPC contig ctg255 (see Figure 3). This result suggests that either FPC contig ctg121 or ctg255 was incorrectly placed. In the same way, each half of the map contigs OMcontigs_1, 9, 12, 16, and 17 was also realigned, and this led to improved placements on the iMap. In all, these findings suggest that the current assigned locations of some FPC contigs (ctg166, ctg172, ctg180, ctg183, ctg197, ctg331, ctg332, ctg377 and ctg378) should be reevaluated.
Figure 3. Optical contig placements on iMap using BACop.
iMap chromosomes are numbered tracks showing the locations of FPC contigs and their subsidiary BACs. BACs (small boxes) are colored keyed according to their sequencing status: magenta [HTGS_FULLTOP - 2×384 paired-end attempts (6× coverage), completed shotgun phase, initial assembly]; lime [HTGS_PREFIN - completed automated improvement phase (AutoFinish)]; cyan [HTGS_ACTIVEFIN - active work being done by a finisher], yellow [HTGS_IMPROVED - finished sequence in gene regions; improved regions will be indicated, once order and orientation of improved segments are confirmed; a comment will be added to indicate this], and black [BACs with no usable or complete SwaI fragments]. The inset shows a zoomed view of a region (ctg146–153) on Chr 3. The blue tracks show optical map contigs anchored to the iMap by BACop. Optical contig identifiers are lettered in blue; pink lettering indicates that an OMcontig was split into two or more pieces for optimizing alignments. Black lettering and a “+,” indicate that two or more optical map contigs were joined. Vertical grey lines show placements of BACs onto optical map contigs. OM = optical map.
We assessed the accuracy of BACop by analyzing the expected placement of 15 telomeric optical contigs onto the ends of chromosomes on the iMap. Figure 3 indeed shows their placement at chromosome ends: OMcontigs_23, 28, 31, 35, 38, 39, 47, and 61 are respectively anchored on the rightmost ends of Chrs 9, 5, 7, 8, 4, 2, 3, and 1. OMcontigs_7, 13, 16, 20, and 21 are respectively anchored on the leftmost ends of Chrs 5, 3, 8, 7, and 10. Also, OMcontig_51 is anchored on FPC contigs ctg368–370 without covering the leftmost end of FPC contig ctg367 on Chr 9, and OMcontig_8 is anchored on FPC contigs ctg405–420 without covering the rightmost end of FPC contig ctg421 on Chr 10. These results suggest that FPC contigs ctg367 and ctg421 should be placed elsewhere, since OMcontigs_51 and 8 have contig “edges” that may represent telomeric regions. The telomeric portion of OMcontig_28 is anchored on FPC contig ctg255 at the rightmost end of Chr 5, and the other portion of OMcontig_28 is anchored on FPC contig ctg121, which is placed on the iMap Chr 3 pericentromeric region. Our findings here indicate that FPC contig ctg121 probably should be joined with ctg255 on Chr 5.
Optical versus sequence alignments identify discordances
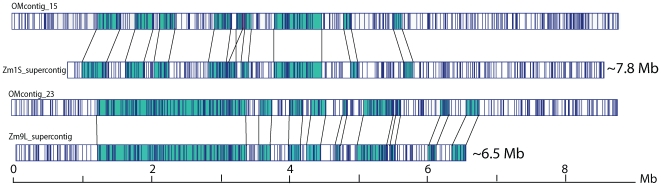
We evaluated the quality of available and ongoing maize sequence assemblies by comparing optical contigs completely spanning large “supercontigs” (pseudomolecules) from Chrs 1, 3 and 9 [54] (Figure 4 and data not shown). Our alignments show 9 map segments in common, spanning 2.29 Mb (29.37%), between OMcontig_15 and the Zm1S_supercontig (Chr 1) in silico restriction map, and 9 in common between OMcontig_23 and Zm9L_supercontig (Chr 9) covering 3.62 Mb (54.85%). However, the Chr 3 finished supercontigs, corresponding to GenBank EF517601 and EF517600 [17], respectively, showed perfect alignment within OMcontigs_13 and 46, demonstrating the efficacy of our approach (data not shown). The lack of comprehensive alignment between the optical and the in silico maps for Chrs 1 and 9 pseudomolecules is not surprising because most of the sequenced BACs are in phase 1 assembly, awaiting the ordering and orienting of their associated sequence contigs. Accordingly, gaps of unknown size remain both within and between these nascent sequence assemblies. Based on these alignments of optical contigs vs. pseudomolecules, we characterized many of these gaps and identified issues with orientation. The assembly of the Zm9L_supercontig appears to be superior to that of the Zm1S_supercontig. This view is further buttressed by the higher proportion of phase 2 BAC sequences (28/56) in the Zm9L_supercontig than in the Zm1S_supercontig (14/60).
Figure 4. Comparative alignment of optical and pseudomolecule maps.
In silico restriction maps of pseudomolecules (Zm1S_supercontig and Zm9L_supercontig) were found to align (Materials and Methods) to optical contigs (OMcontig_15 and 23). This allowed the identification of common and discordant regions. SwaI restriction sites are depicted by vertical lines. Regions of the optical contig and the pseudomolecule that align are teal colored, and the aligned regions are pointed to with black connecting lines.
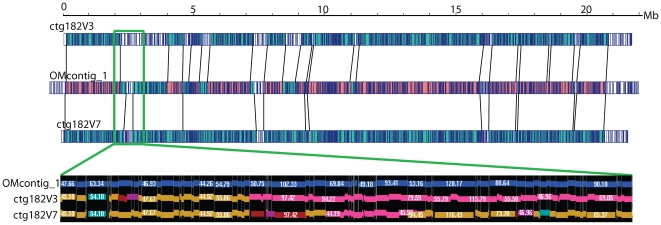
The process of constructing a large sequence pseudomolecule is an iterative one, drawing support for provisional assembly from many sources that guide the serial generation of hypothetical builds and their subsequent validation. As such, we performed a series of optical vs. sequence contig alignments that tracked, guided, and validated the ongoing sequence finishing efforts of a ∼22 Mb sequence pseudomolecule (FPC ctg182) by the Arizona Genomics Institute (AGI). Figure 5 shows two versions of ctg182, V3 and V7, aligned to the optical contig—OMcontig_1. The earliest sequence contig build (V3) contained 12 segments that aligned (∼74.9%) with the optical contig, totaling 16.30 Mb. Rounds of directed sequence finishing effort led to the construction of the updated build, V7, which addressed discordances. V7 and the optical contig show an increased alignment of 89.6% with 8 larger segments aligning, totaling 19.51 Mb.
Figure 5. Optical maps guide ongoing construction of pseudomolecules.
The quality of a large pseudomolecule (ctg182; ∼21.8 Mb) was successively improved by alignment of provisional builds against OMcontig_1 (optical contig). The increase in aligned regions of ctg182V7 to OMcontig_1 compared to the earlier version, ctg182V3, to the OMcontig demonstrates the improvement in the quality of the build. Red highlighting in OMcontig_1 shows optical contig regions aligning to both pseudomolecule versions. Maps of the ∼1.3 M region boxed in green are shown below, with concordances and discordances illustrated between the optical contig and ctg182V3 and V7 on a per restriction fragment basis. Gold colored fragments (boxes) signify concordance, while other colors signify discordance. Sizes are in kb. Ctg182V3 contains a run of pink fragments, indicating discordance with OMcontig_1, which is partially mediated in ctg182V7 as evidenced by greater alignment (less pink and more gold boxes).
Toward the AGP (B73 RefGen_v1): placement of 435 FPC contig pseudomolecules onto optical contig maps
The assembly of the accessioned golden path (AGP) involved the merging of 435 correctly ordered FPC pseudomolecules (90.2 kb–34.783 Mb; 2,061 Mb total) to span across the entire maize genome, and it is now known as the B73 RefGen_v1 [53],[79]. These FPC contig pseudomolecules are constructed from recent sequencing data (16,848 tiled BACs). We facilitated the construction of the AGP by ordering these 435 FPC pseudomolecules, using their alignment to our optical contigs to place them. We uniquely placed 338 of the 435 FPC pseudomolecules onto optical maps; 16 were placed on two optical map contigs, bridging two optical map contigs. Alignments also revealed two possible FPC chimeras (ctg84 and ctg299; Table S1). The remaining 82 FPC pseudomolecules (∼63 Mb) were not placed on optical contigs due to regions bearing few SwaI sites, or to problems in sequence assembly. Among the 338 uniquely placed FPC pseudomolecules, 65 (∼19%) are either newly placed (33; Table S1; blue rows) or reassigned to amended locations (32; Table S1; yellow rows). Overall, these results demonstrate the utility of a scalable optical map framework for guiding sequence assembly within a complex genomic environment.
Comparing optical maps and B73 RefGen_v1 sequence
The accessioned golden path (AGP)(B73 RefGen_v1) from the Arizona Genomics Institute recently released by the maize genome sequencing consortium comprises 10 chromosome-wide “reference chromosomes” (pseudomolecules), and its assembly was guided by several physical maps, including the optical mapping findings presented here (http://www2.genome.arizona.edu/genomes/maize_contig_quality_table). The B73 RefGen_v1 reference chromosomes represent a unified genomic resource showing chromosome-wide placement of sequence and associated gaps. We provided an independent, optical reference map for this important resource via alignments that comprehensively revealed and sized sequence gaps in FPC pseudomolecules and the B73 RefGen_v1, which compose reference chromosomes. Local alignment reveals AGP assembly errors characterized as novel gaps, extra and/or missing cuts, and fragment sizing errors. In total, 1,102 optical contig segments (strings of contiguous restriction fragments) aligned to the B73 RefGen_v1 reference chromosomes (1,014.49 Mb, or ∼50% of AGP [2,046.35 Mb]; Table 2). The number of optical contig segments that align per chromosome ranges from 74 (Chr 10) to 159 (Chr 1), and the average map segment size is 937.67 kb (Table 2). The total aligned mass per chromosome varies from 64.65 Mb (Chr 8) to 166.15 Mb (Chr 1). The coverage by the aligned map segments for all the maize chromosomes ranges from 37.04% (Chr 8) to 59.15% (Chr 4) and averages 49.58% for all chromosomes (Table 2). Since the construction of the B73 RefGen is still ongoing, we expected that the optical map: B73 RefGen_v1 alignments would reveal a high level of discordance and an attenuated rate of optical contig alignment. A total of 4,465 discordances are identified (Table S2). These findings include 564 loci with extra sequence data, 829 revealing novel gaps or missing sequences, 2,348 misassemblies, 478 additional SwaI restriction sites, and 246 missing SwaI restriction sites.
Table 2. Statistics for optical map alignments against the in silico maps of the B73 RefGen_v1.
| Chr. No. | Ref. Chr. Size (Mb) | No. of Aligned Map Segments | Ave. Map Segment Size (kb) | Total Aligned Map Segment Size (Mb) | % Coverage |
| 1 | 300.24 | 159 | 1044.97 | 166.15 | 55.34 |
| 2 | 234.75 | 121 | 1258.93 | 125.33 | 53.39 |
| 3 | 230.56 | 132 | 873.33 | 115.28 | 50.00 |
| 4 | 247.10 | 109 | 1340.83 | 146.15 | 59.15 |
| 5 | 216.92 | 134 | 702.39 | 94.12 | 43.39 |
| 6 | 169.25 | 92 | 839.02 | 77.19 | 45.61 |
| 7 | 170.97 | 88 | 944.21 | 83.09 | 48.60 |
| 8 | 174.52 | 103 | 627.67 | 64.65 | 37.04 |
| 9 | 152.35 | 90 | 836.33 | 75.27 | 49.41 |
| 10 | 149.69 | 74 | 909.05 | 67.27 | 44.94 |
| Total/ Ave. | 2046.35 | 1102 | 937.67 | 1014.49 | 49.58 |
* Note: Chr. = chromosome, Ref. Chr. = reference chromosome, Ave. = average.
Gaps between adjacent FPC contigs were sized by alignments of optical contigs that span across them. Accordingly, gap size is determined by comparing optical map and B73 RefGen_v1 coordinates across a gap formed between neighboring FPC contigs in the B73 RefGen_v1 reference chromosomes. The FPC contigs do not continuously and seamlessly align to optical contigs since they are constructed from unfinished BACs (Figure 3 shows optical alignments to FPC contigs). Thus we estimated B73 RefGen_v1 gap sizes by considering the pair of coordinates on an aligned optical contig that most closely flank the spanned FPC gap (Figure S1). More precisely: Gap (kb) = [|(right optical coordinate)−(left optical coordinate)|−|(right sequence coordinate)−(left sequence coordinate)|]/1000. In this way, we characterized 263 gaps (Table S3) comprising 44 “negative gaps” (false B73 RefGen_v1 gaps, or novel sequence) and 219 “positive gaps” (confirmed FPC gaps, or unaccounted sequence). These 263 gaps were called taking the optical mapping sizing error per restriction fragment into consideration, which is typically +/−5% [80]. However, optical sizing errors can accrue in a complex way across long genomic regions that are spanned by summing consecutive restriction fragments [81]–[82]. As such, 169 of the 263 gap calls were conservatively made when the AGP and optical alignments differences were ≥10%, and the remainder was called below this threshold. Here differences <10% indicate the presence of gaps that were called with less confidence, but their tabulation provides considered targets for sequence bridging and filling. In all, 155 gaps were bridged by optical contigs (covering 36.59 Mb of gaps), and an extra 2.09 Mb of AGP pseudomolecule sequence was identified.
Discussion
An optical map was created that spans across ∼91% of the maize (Zea mays L.) B73 inbred line (PI 550473) genome, which is a parent of the IBM mapping population. 66 optical contigs are included in this map representing 2,103.93 Mb of the maize genome decorated by 91,453 ordered SwaI restriction sites with accurate physical distances between these sites. On average, there is a SwaI site every 23 kb across the genome, and this restriction recognition sequence “marker” density is far greater than those on genetic (∼6,000 markers) and FPC (∼24,000 markers) maps [37] (http://maize-mapping.plantgenomics.iastate.edu/). Because the optical data format is a high-resolution ordered restriction map (SwaI), we were able to anchor and orient FPC-sequence contigs (http://www2.genome.arizona.edu/genomes/maize_contig_quality_table) onto this scaffold. While the immediate utility of the maize optical reference map is as an independent reference for sequence finishing and closing gaps, it will also drive comparative studies for unraveling complex patterns of structural variation as additional inbred lines and cultivars are mapped. Here the optical reference map would serve as a scaffold for future map assemblies enabling rapid discernment of genomic architecture. In this regard, optical mapping may be unique since large ∼500 kb molecules are directly mapped, and this advantage supports scalable genome analysis spanning from a restriction site to multi-megabase-sized regions.
The de novo approach that we used to construct the maize optical reference map ensures that it a unique, purely independent resource for sequence assembly and validation. (The ∼2.1 Gb map constructed de novo represents the largest created using single, genomic DNA molecules.) This physical map is free from common cloning and PCR artifacts, since individual genomic molecules are directly analyzed. These advantages are demonstrated by our comprehensive analysis of several pseudomolecules (Figure 4 and Figure 5), both published and ongoing, as well as B73 RefGen_v1 reference chromosomes (Table S3) spanning the entire maize genome.
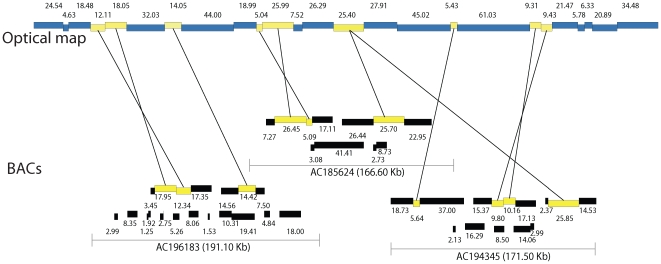
Our development of a new algorithm, named BACop (Materials and Methods; Figure 2, Figure 3, and Figure 6), greatly facilitated our ability to analyze and contribute to ongoing sequencing efforts. BACop specifically addressed issues of aligning nascent sequence builds of BAC clones, harboring multiple unordered and unoriented contigs (averaging 11 per BAC), against the optical reference map. Here, BACop was able to link optical contig maps to many unfinished BAC sequences already placed on the FPC map [37], and to presciently orient and order optical findings across all 10 maize chromosomes. Furthermore, BACop enabled the placement of 60 of the 66 optical contigs onto iMap and identified 12 FPC contigs whose current placement on the iMap requires reevaluation. The calls on 11 out of the 12 FPC contigs identified by BACop for replacements on the iMap (ctg121, ctg166, ctg172, ctg180, ctg183, ctg197, ctg332, ctg333, ctg367, ctg377 and ctg378) were also supported by comparative analysis of optical maps and in silico maps of the FPC contig sequence pseudomolecules (Table S1). The additional FPC contig identified for replacement by BACop (ctg421) was supported by other sequence markers indicating a new placement abutting ctg90 on chromosome 2 [79]. This BACop algorithm, in combination with optical map data, can also order and orient nascent sequence assemblies. As shown in Figure 6, many of the unordered and unoriented BAC subcontigs for several clones are nicely placed onto an optical map. Accordingly, BACop provides a useful tool for guiding the ongoing finishing of individual BACs.
Figure 6. Alignment of unfinished BAC sequence contigs to optical contig maps.
Matching restriction fragments in the BAC sub-contigs and the optical contig map are indicated by yellow boxes connected by lines; numbers show fragment size (kb). These alignments illustrate how BAC sequence contigs can be ordered and oriented using optical map alignments.
Given the abundance of maize repetitive sequence, restriction maps directly constructed from ∼500 kb genomic molecules offer many advantages for spanning and structurally characterizing heterochromatic regions. This advantage is evidenced by the long contigs (3.64 Mb to 100.76 Mb) within the optical reference map, averaging 31.88 Mb in length. In comparison, the May 2006 maize FPC map comprises 721 FPC contigs averaging ∼2.98 Mb in length with a total mass of 2,150 Mb (300/721, or 163.7 Mb, are unassigned) [37]. Long optical contigs offer unique benefits especially when they span across genomic regions sparsely populated by markers, enabling structural insights to be drawn in these regions. In part, these advantages have characterized gaps and sequence misassemblies and reordered 19% of the FPC pseudomolecules (Table S1), which was persisted into the current B73 RefGen_v1 sequence for the maize genome.
Maize centromeres have been mapped to regions with flanking molecular markers using many different approaches [83]–[88]. However, for some maize chromosomes such as Chrs 1, 3, and 6, the proposed centromeric locations differ among different mapping methods; while for other chromosomes - Chrs 2, 4, 9, and 10 - there is a consensus running across different mapping techniques [84]. Recently, maize centromeres were located on the B73 RefGen_v1 sequence using centromeric markers [53] derived from numerous sources: transposon display, repeat junction, centromere repeat, and chromatin immunoprecipitation data [53]. Accordingly, we located centromeric loci assigned to Chrs 1, 2, 5, 6, 8, 9, and 10 around gaps in the optical contigs: [CEN1 (OMcontig_3 and 69); CEN2 (OMcontig_2 and 41); CEN5 (OMcontig_22 and 42); CEN6 (OMcontig_65 and 36); CEN8 (OMcontig_16 and 4); CEN 9 (OMcontig_55 and 17); CEN10 (OMcontig_49 and 17)] (data not shown). Unfortunately, the flanking contigs did not fully span any centromeric regions; however, these optical contigs did structurally characterize several pericentric regions.
Although we have demonstrated here that optical mapping offers numerous benefits for physical mapping and genome sequence assembly, its utility would be appreciably extended when combined with next generation sequencing. Genome analysis approaches are now rapidly evolving and tracking the increasingly cost-effective capabilities offered by next generation sequencing. Next generation sequencing approaches using single molecule libraries are now tackling the analysis of complex genomes [89]–[90], but they do not offer data sets competent for de novo assembly because of modest read lengths and errors. As such, new sequencing strategies must be developed for wheat and other complex crop genomes that effectively seize the new opportunities enabled by next generation sequencing. To this end, we propose the proactive use of optical mapping data for sequence assembly [91]. The combination of long-range (optical) and nucleotide-level (next generation) data sets, both generated directly from genomic molecules, may prove to be a cost-effective approach - especially when new algorithms are developed that intimately comingle both data sets during the assembly process.
Materials and Methods
Seed germination and DNA preparation
Maize kernels (inbred line B73, PI550473), obtained from the USDA-Agriculture Research Service North Central Regional Plant Introduction Station (Iowa State University, Ames, IA 50011-1170), were washed in 10% Clorox bleach for 10 min, rinsed in sterile water (3×, ∼3 min per wash), and germinated on moistened brown paper towels in a dark, moist chamber at 30°C for 12 days. Residual ungerminated seeds were removed from maize sprouts prior to nuclei isolation. The procedures for isolation of nuclei and storage have been described previously [75]. Prior to use, isolated nuclei were washed 2× with fresh Dulbecco's PBS (1.54 mM KH2PO4, 155.17 mM NaCl, 2.71 mM Na2HPO4, pH 7.2) to remove glycerol. Rapid DNA concentration assays were conducted by lysing small aliquots of nuclei in TE (10 mM Tris-Cl, pH 8.0, 1 mM EDTA) with 1 mg/ml proteinase K, and adenovirus DNA added at 25 pg/µl (internal sizing standard; Invitrogen, Carlsbad, CA), followed by mounting, restriction digestion, staining and imaging as previously described. Appropriate dilutions for mapping (optimized to minimize molecular crossovers) were made by adjusting the amount of isolated nuclei in the lysing solution (TE with 1 mg/ml proteinase K, 25 pg/µl adenovirus DNA in TE), by slowly pipetting up and down several times using a wide-bore pipette tip; samples were incubated at 65°C for 1 hr and at 37°C overnight. Samples were mounted onto optical mapping surfaces and imaged by fluorescence microscopy to assess DNA integrity and concentration of both genomic and reference standard DNA molecules.
Surface preparation
Surface preparation was done as previously described [71]–[72]. Briefly, glass cover slips (22×22 mm, Fisher's Finest) were cleaned by boiling in Nano-Strip (Cyantek Corp., Freemont, CA), acidified by boiling in concentrated HCl, extensively rinsed with high purity water and ethanol under sonication, and derivatized using trimethyl and vinyl silanes to confer a positive charge and the means to crosslink the acrylamide overlay to the surface. Surfaces were evaluated by mounting lambda DASH II bacteriophage DNA (Stratagene, La Jolla, CA) and digesting them with 40 units of SwaI, diluted in 100 µL of digestion buffer containing 0.02% Triton X-100 at room temperature to determine the optimal digestion time (30 min to 2.5 hrs).
DNA mapping, image acquisition, and processing
Genomic DNA molecules (∼400–500 kb) premixed with the adenovirus DNA sizing standards were deposited as stripes on derivatized glass surfaces using a silastic microchannel system [92]. A fully automated image acquisition and processing system collected data and compiled large files consisting of an ordered restriction map for each genomic DNA molecule. All microscope and camera functionalities and machine vision processes are fully automated and controlled by computer software. Detailed procedures were previously described [74]–[76],[92].
Map assembly and cluster computing
With a raw map data set of >2 million maps, a divide and conquer strategy for optical map assembly was needed to deal with the severe computational load through parallel processing. We previously used this approach for the assembly of genome maps spanning the rice as well as the Leishmania major genomes [74]–[75]. Briefly, the map data set was divided into smaller sub-data sets (∼30,000 single molecule optical maps) allowing efficient parallel assembly [93]–[96], over 2–3 days, without taxing computer memory limits. The consensus maps from all contig assemblies were reassembled together for identifying redundant contigs and for merging overlapping optical consensus maps. After this reassembly process, a unique set of optical consensus maps was identified as “seed” maps for initiating iterative assembly. Iterative assembly consists of cycles of pairwise alignment [78] of the entire map data set against seed maps, followed by the contiging of these aligned single molecular optical maps for extending and refining seed maps in each subsequent cycle. Cycles of iterative assembly broaden and increase the coverage depth of nascent contigs.
Consensus maps are then stripped from the newly formed contigs as updated seed maps after further processing. The pairwise alignment phase extracted multiple high-scoring alignments based on the efficient linear scaling approach of Huang and Miller [97], and the confidence scores (p-values) were generated using an approach similar to that used by Waterman and Vingron [98]. Updated consensus maps were assembled to identify redundancy and to merge overlapping consensus maps. This process identified seed maps for the next iteration, and this iteration process was repeated typically more than ten times until the optical map contigs no longer grew. Large contigs (>10 Mb in breadth) also present computational challenges. For these contigs, iterative assembly considers and augments only the terminal 40 restriction fragments.
Development of a new algorithm, BACop, for anchoring optical map contigs onto BAC sequences within iMap
About 85% of the maize genome comprises extensive families of repetitive sequences. Consequently, multiple contigs emerge from the sequence assembly of a single BAC, which are also unordered and unoriented. In order to integrate our optical map with the iMap, we developed a new algorithm —“BACop” — that utilizes unfinished BAC sequence data. The algorithm for anchoring the optical maps on the maize genetic-physical (FPC) map precedes in four distinct steps: i) matching restriction fragments between the optical map and the in silico restriction fragments from the sequence contigs of the BACs, ii) determining locations of all the BAC sequence contigs along the optical map, iii) anchoring the BACs on the optical maps, and iv) filtering and combining the alignments of BACs in the FPC map and optical map to find the most significant ones (Figure 2). The first step compares individual restriction fragment sizes from the optical map contigs with the in silico restriction fragments of the sequence contigs of the BACs. A fragment from the optical map assembly of size X and an in silico restriction fragment of size Y match if |X−Y|/σ√Y< = k, for parameters σ and k based on the statistical model developed by Valouev et al. 2006 [78]. Once the matching fragments have been determined, the BACs are located on the optical map assembly by examining each BAC's in silico restriction fragments. Approximate locations are determined by a filtration method that selects candidate regions on the optical map assembly that a BAC can align to, based on the matching fragment density. The approximate locations are further screened to produce a feasible alignment of a BAC's restriction fragments to the restriction fragments on the optical map assembly. Since each BAC is shotgun sequenced, multiple sequence contigs can result since the orientation and order of sequences are unknown. A feasible alignment must preserve the order of the in silico restriction fragments from within the same sequence contig but is allowed to match fragments from different sequence contigs in any order and orientation.
A match graph is constructed from a candidate region on the optical map assembly that the BAC can align to using the matching restriction fragments between the two as nodes within the graph. A traversal through the match graph induces an alignment of the BAC to the optical map assembly from the nodes representing matching fragments along the traversed path. The graph traversal resembles a branch and bound algorithm that exhaustively enumerates all feasible alignments and selects the best one. After all possible locations of the BACs are determined, the optical contig is then aligned to the FPC map. The FPC map gives the relative positions of the BACs within each FPC contig as well as the positions and order of the FPC contigs with respect to each other. Dynamic programming is used to align the optical map contigs to the FPC contigs by scoring for matching BACs along the optical contig that respect the order and location of BACs along the FPC map. A scoring scheme that weighs for higher quality BACs based on sequencing status and for BACs with greater fragment density is used. A fudge factor is applied when examining the locations of the BACs specified by the FPC map since they are approximate. Gaps between FPC contigs are specified using lower and upper bounds. The alignment is evaluated based on the number of BACs that are scored within the alignment region to remove spurious alignments according to a set threshold. The threshold is adjusted to allow for alignments in regions where there is sparse sequence data resulting in a lower number of usable BACs to align to. All of the alignments made with different thresholds are then collected, and the best alignments are selected according to coverage of the optical map assembly and number of aligned BACs.
Supporting Information
Using optical contigs to estimate sizes of gaps between adjacent FPC contigs. (A) Restriction map view (box = restriction fragment) of FPC contigs aligned to the AGP Chr 6 pseudomolecule; the gap is highlighted. (B) Cartoon showing the alignments of FPC contigs ct280 and ctg281 to Chr6 pseudomolecule. A gap of unknown size is illustrated, with green lines showing alignment to the pseudomolecule. Dashed lines show alignments of ct280 and ctg281 against OMcontig_6 (gold track) and a large gap. (C) Restriction map view of OMcontig_6, locating the large gap within the grayed restriction fragments. FPC gap sizes are calculated as: Gap (kb) = [|(right optical coordinate)−(left optical coordinate)|−|(right sequence coordinate)−(left sequence coordinate)|]/1000.
(0.30 MB PDF)
Ordering FPC contig sequence pseudomolecules based on the map alignments between optical maps and the in silico maps of the FPC contig sequence pseudomolecules.
(0.10 MB PDF)
Discordances in the well-aligned map segments between optical maps and the in silico maps of the B73 RefGen_v1 reference chromosomes.
(0.25 MB PDF)
FPC gap estimations based on map alignments between optical maps and the in silico maps of FPC contig sequence pseudomolecules and B73 RefGen_v1 reference chromosomes.
(0.12 MB PDF)
Acknowledgments
We thank Brent Kronmiller for expert assistance with the Chr 3 supercontigs prior to publication, the USDA–ARS North Central Regional Plant Introduction Station (NCRPIS), John Doebley for providing maize B73 (PI 550473) germplasms for this project, and David Lu for assisting with map alignments. We also thank the UW–Madison Condor computing group for computational resources.
Footnotes
The authors have declared that no competing interests exist.
Funding provided by NSF DBI-0501818 and NHGRI R01 HG000225 (DCS). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Rayburn AL, Biradar DP, Bullock DG, McMurphy LM. Nuclear DNA content in F-1 hybrids of maize. Heredity. 1993;70:294–300. [Google Scholar]
- 2.Gaut BS, Doebley JF. DNA sequence evidence for the segmental allotetraploid origin of maize. Proc Natl Acad Sci U S A. 1997;94:6809–6814. doi: 10.1073/pnas.94.13.6809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Swigonova Z, Lai J, Ma J, Ramakrishna W, Llaca V, et al. On the tetraploid origin of the maize genome. Comp Funct Genomics. 2004;5:281–284. doi: 10.1002/cfg.395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gaut BS. Patterns of chromosomal duplication in maize and their implications for comparative maps of the grasses. Genome Res. 2001;11:55–66. doi: 10.1101/gr.160601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gaut BS, Le Thierry d'Ennequin M, Peek AS, Sawkins MC. Maize as a model for the evolution of plant nuclear genomes. Proc Natl Acad Sci U S A. 2000;97:7008–7015. doi: 10.1073/pnas.97.13.7008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ilic K, SanMiguel PJ, Bennetzen JL. A complex history of rearrangement in an orthologous region of the maize, sorghum, and rice genomes. Proc Natl Acad Sci U S A. 2003;100:12265–12270. doi: 10.1073/pnas.1434476100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lai J, Ma J, Swigonova Z, Ramakrishna W, Linton E, et al. Gene loss and movement in the maize genome. Genome Res. 2004;14:1924–1931. doi: 10.1101/gr.2701104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Langham RJ, Walsh J, Dunn M, Ko C, Goff SA, et al. Genomic duplication, fractionation and the origin of regulatory novelty. Genetics. 2004;166:935–945. doi: 10.1534/genetics.166.2.935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.SanMiguel P, Gaut BS, Tikhonov A, Nakajima Y, Bennetzen JL. The paleontology of intergene retrotransposons of maize. Nat Genet. 1998;20:43–45. doi: 10.1038/1695. [DOI] [PubMed] [Google Scholar]
- 10.Bennetzen JL, SanMiguel P, Chen M, Tikhonov A, Francki M, et al. Grass genomes. Proc Natl Acad Sci U S A. 1998;95:1975–1978. doi: 10.1073/pnas.95.5.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.SanMiguel P, Tikhonov A, Jin YK, Motchoulskaia N, Zakharov D, et al. Nested retrotransposons in the intergenic regions of the maize genome. Science. 1996;274:765–768. doi: 10.1126/science.274.5288.765. [DOI] [PubMed] [Google Scholar]
- 12.Rabinowicz PD, Bennetzen JL. The maize genome as a model for efficient sequence analysis of large plant genomes. Curr Opin Plant Biol. 2006;9:149–156. doi: 10.1016/j.pbi.2006.01.015. [DOI] [PubMed] [Google Scholar]
- 13.Ananiev EV, Phillips RL, Rines HW. Complex structure of knob DNA on maize chromosome 9. Retrotransposon invasion into heterochromatin. Genetics. 1998;149:2025–2037. doi: 10.1093/genetics/149.4.2025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ananiev EV, Phillips RL, Rines HW. A knob-associated tandem repeat in maize capable of forming fold-back DNA segments: are chromosome knobs megatransposons? Proc Natl Acad Sci U S A. 1998;95:10785–10790. doi: 10.1073/pnas.95.18.10785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Meyers BC, Tingey SV, Morgante M. Abundance, distribution, and transcriptional activity of repetitive elements in the maize genome. Genome Res. 2001;11:1660–1676. doi: 10.1101/gr.188201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kronmiller BA, Wise RP. TEnest: automated chronological annotation and visualization of nested plant transposable elements. Plant Physiol. 2008;146:45–59. doi: 10.1104/pp.107.110353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kronmiller BA, Wise RP. Computational finishing of large sequence contigs reveals interspersed nested repeats and gene islands in the rf1-associated region of maize. Plant Physiology. 2009;150 doi: 10.1104/pp.109.143370. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Emerson RA, Beadle GW, Fraser AC. A summary of linkage studies in maize. Cornell Univ Agric Exp Stn Memoir. 1935;180:1–83. [Google Scholar]
- 19.Fu Y, Wen TJ, Ronin YI, Chen HD, Guo L, et al. Genetic dissection of intermated recombinant inbred lines using a new genetic map of maize. Genetics. 2006;174:1671–1683. doi: 10.1534/genetics.106.060376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fang Z, Polacco M, Chen S, Schroeder S, Hancock D, et al. cMap: the comparative genetic map viewer. Bioinformatics. 2003;19:416–417. doi: 10.1093/bioinformatics/btg012. [DOI] [PubMed] [Google Scholar]
- 21.Roman H, Ullstrup AJ. The use of B-A translocations to locate genes in maize. Agron J. 1951;43:450–454. [Google Scholar]
- 22.Helentjaris T, Weber DF, Wright S. Use of monosomics to map cloned DNA fragments in maize. Proc Natl Acad Sci U S A. 1986;83:6035–6039. doi: 10.1073/pnas.83.16.6035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Burr B, Burr FA, Thompson KH, Albertson MC, Stuber CW. Gene mapping with recombinant inbreds in maize. Genetics. 1988;118:519–526. doi: 10.1093/genetics/118.3.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Coe EH, Hoisington DA, Neuffer MG. Linkage map of Corn (maize) (Zea mays L.). Maize Genet Coop Newslett. 1987;61:116–147. [Google Scholar]
- 25.Gardiner JM, Coe EH, Melia-Hancock S, Hoisington DA, Chao S. Development of a core RFLP map in maize using an immortalized F2 population. Genetics. 1993;134:917–930. doi: 10.1093/genetics/134.3.917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Weber D, Helentjaris T. Mapping RFLP loci in maize using B-A translocations. Genetics. 1989;121:583–590. doi: 10.1093/genetics/121.3.583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Beavis WD, Lee M, Grant D, Hallaur AR. The influence of random mating on recombination among RFLP loci. Maize Gen Coop Newsl. 1992;66:52–53. [Google Scholar]
- 28.Davis GL, McMullen MD, Baysdorfer C, Musket T, Grant D, et al. A maize map standard with sequenced core markers, grass genome reference points and 932 expressed sequence tagged sites (ESTs) in a 1736-locus map. Genetics. 1999;152:1137–1172. doi: 10.1093/genetics/152.3.1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Veldboom LR, Lee M. Genetic mapping of quantitative trait loci in maize in stress and nonstress environments: II. Plant height and flowering. Crop Science. 1996;36:1320–1327. [Google Scholar]
- 30.Taramino G, Tingey S. Simple sequence repeats for germplasm analysis and mapping in maize. Genome. 1996;39:277–287. doi: 10.1139/g96-038. [DOI] [PubMed] [Google Scholar]
- 31.Lee M. Comparative genetic and QTL mapping in sorghum and maize. Symp Soc Exp Biol. 1996;50:31–38. [PubMed] [Google Scholar]
- 32.Causse M, Santoni S, Damerval C, Maurice A, Charcosset A, et al. A composite map of expressed sequences in maize. Genome. 1996;39:418–432. doi: 10.1139/g96-053. [DOI] [PubMed] [Google Scholar]
- 33.Senior ML, Heun M. Mapping maize microsatellites and polymerase chain reaction confirmation of the targeted repeats using a CT primer. Genome. 1993;36:884–889. doi: 10.1139/g93-116. [DOI] [PubMed] [Google Scholar]
- 34.Edwards KJ, Thompson H, Edwards D, de Saizieu A, Sparks C, et al. Construction and characterisation of a yeast artificial chromosome library containing three haploid maize genome equivalents. Plant Mol Biol. 1992;19:299–308. doi: 10.1007/BF00027351. [DOI] [PubMed] [Google Scholar]
- 35.Cone KC, McMullen MD, Bi IV, Davis GL, Yim YS, et al. Genetic, physical, and informatics resources for maize. On the road to an integrated map. Plant Physiol. 2002;130:1598–1605. doi: 10.1104/pp.012245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Coe E, Cone K, McMullen M, Chen SS, Davis G, et al. Access to the maize genome: an integrated physical and genetic map. Plant Physiol. 2002;128:9–12. doi: 10.1104/pp.128.1.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wei F, Coe E, Nelson W, Bharti AK, Engler F, et al. Physical and Genetic Structure of the Maize Genome Reflects Its Complex Evolutionary History. PLoS Genet. 2007;3:e123. doi: 10.1371/journal.pgen.0030123. doi: 10.1371/journal.pgen.0030123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sharopova N, McMullen MD, Schultz L, Schroeder S, Sanchez-Villeda H, et al. Development and mapping of SSR markers for maize. Plant Mol Biol. 2002;48:463–481. doi: 10.1023/a:1014868625533. [DOI] [PubMed] [Google Scholar]
- 39.Soderlund C, Longden I, Mott R. FPC: A system for building contigs from restriction fingerprinted clones. Computer Applications in the Biosciences. 1997 doi: 10.1093/bioinformatics/13.5.523. [DOI] [PubMed] [Google Scholar]
- 40.Haldi M, Perrot V, Saumier M, Desai T, Cohen D, et al. Large human YACs constructed in a rad52 strain show a reduced rate of chimerism. Genomics. 1994;24:478–484. doi: 10.1006/geno.1994.1656. [DOI] [PubMed] [Google Scholar]
- 41.Kim UJ, Shizuya H, de Jong PJ, Birren B, Simon MI. Stable propagation of cosmid sized human DNA inserts in an F factor based vector. Nucleic Acids Res. 1992;20:1083–1085. doi: 10.1093/nar/20.5.1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yim YS, Davis GL, Duru NA, Musket TA, Linton EW, et al. Characterization of three maize bacterial artificial chromosome libraries toward anchoring of the physical map to the genetic map using high-density bacterial artificial chromosome filter hybridization. Plant Physiol. 2002;130:1686–1696. doi: 10.1104/pp.013474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tomkins JP, Davis G, Main D, Yim Y, Duru N, et al. Construction and characterization of a deep-coverage bacterial artificial chromosome library for maize. Crop Science. 2002;42:928–933. [Google Scholar]
- 44.Gardiner J, Schroeder S, Polacco ML, Sanchez-Villeda H, Fang Z, et al. Anchoring 9,371 maize expressed sequence tagged unigenes to the bacterial artificial chromosome contig map by two-dimensional overgo hybridization. Plant Physiol. 2004;134:1317–1326. doi: 10.1104/pp.103.034538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fang Z, Cone K, Sanchez-Villeda H, Polacco M, McMullen M, et al. iMap: a database-driven utility to integrate and access the genetic and physical maps of maize. Bioinformatics. 2003;19:2105–2111. doi: 10.1093/bioinformatics/btg289. [DOI] [PubMed] [Google Scholar]
- 46.Soderlund C, Humphray S, Dunham A, French L. Contigs built with fingerprints, markers, and FPC V4.7. Genome Res. 2000;10:1772–1787. doi: 10.1101/gr.gr-1375r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nelson WM, Bharti AK, Butler E, Wei F, Fuks G, et al. Whole-genome validation of high-information-content fingerprinting. Plant Physiol. 2005;139:27–38. doi: 10.1104/pp.105.061978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Messing J, Bharti AK, Karlowski WM, Gundlach H, Kim HR, et al. Sequence composition and genome organization of maize. Proc Natl Acad Sci U S A. 2004;101:14349–14354. doi: 10.1073/pnas.0406163101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang Q, Dooner HK. Remarkable variation in maize genome structure inferred from haplotype diversity at the bz locus. Proc Natl Acad Sci U S A. 2006;103:17644–17649. doi: 10.1073/pnas.0603080103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Song R, Messing J. Gene expression of a gene family in maize based on noncollinear haplotypes. Proc Natl Acad Sci U S A. 2003;100:9055–9060. doi: 10.1073/pnas.1032999100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yao H, Zhou Q, Li J, Smith H, Yandeau M, et al. Molecular characterization of meiotic recombination across the 140-kb multigenic a1-sh2 interval of maize. Proc Natl Acad Sci U S A. 2002;99:6157–6162. doi: 10.1073/pnas.082562199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fu H, Dooner HK. Intraspecific violation of genetic colinearity and its implications in maize. Proc Natl Acad Sci U S A. 2002;99:9573–9578. doi: 10.1073/pnas.132259199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schnable PS, Ware D, Fulton RS, Stein JC, Wei F, et al. B73 maize genome: complexity, diversity and dynamics. Science 326. 2009 doi: 10.1126/science.1178534. doi:1126/science. 1178534. [DOI] [PubMed] [Google Scholar]
- 54.Bruggmann R, Bharti AK, Gundlach H, Lai J, Young S, et al. Uneven chromosome contraction and expansion in the maize genome. Genome Res. 2006;16:1241–1251. doi: 10.1101/gr.5338906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Haberer G, Young S, Bharti AK, Gundlach H, Raymond C, et al. Structure and architecture of the maize genome. Plant Physiol. 2005;139:1612–1624. doi: 10.1104/pp.105.068718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Emberton J, Ma J, Yuan Y, SanMiguel P, Bennetzen JL. Gene enrichment in maize with hypomethylated partial restriction (HMPR) libraries. Genome Res. 2005;15:1441–1446. doi: 10.1101/gr.3362105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Barbazuk WB, Bedell JA, Rabinowicz PD. Reduced representation sequencing: a success in maize and a promise for other plant genomes. Bioessays. 2005;27:839–848. doi: 10.1002/bies.20262. [DOI] [PubMed] [Google Scholar]
- 58.Whitelaw CA, Barbazuk WB, Pertea G, Chan AP, Cheung F, et al. Enrichment of gene-coding sequences in maize by genome filtration. Science. 2003;302:2118–2120. doi: 10.1126/science.1090047. [DOI] [PubMed] [Google Scholar]
- 59.Palmer LE, Rabinowicz PD, O'Shaughnessy AL, Balija VS, Nascimento LU, et al. Maize genome sequencing by methylation filtration. Science. 2003;302:2115–2117. doi: 10.1126/science.1091265. [DOI] [PubMed] [Google Scholar]
- 60.Yuan Y, SanMiguel PJ, Bennetzen JL. High-Cot sequence analysis of the maize genome. Plant J. 2003;34:249–255. doi: 10.1046/j.1365-313x.2003.01716.x. [DOI] [PubMed] [Google Scholar]
- 61.Springer NM, Xu X, Barbazuk WB. Utility of different gene enrichment approaches toward identifying and sequencing the maize gene space. Plant Physiol. 2004;136:3023–3033. doi: 10.1104/pp.104.043323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Fernandes J, Brendel V, Gai X, Lal S, Chandler VL, et al. Comparison of RNA expression profiles based on maize expressed sequence tag frequency analysis and micro-array hybridization. Plant Physiol. 2002;128:896–910. doi: 10.1104/pp.010681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Rabinowicz PD, Schutz K, Dedhia N, Yordan C, Parnell LD, et al. Differential methylation of genes and retrotransposons facilitates shotgun sequencing of the maize genome. Nat Genet. 1999;23:305–308. doi: 10.1038/15479. [DOI] [PubMed] [Google Scholar]
- 64.Okagaki RJ, Phillips RL. Maize DNA-sequencing strategies and genome organization. Genome Biol. 2004;5:223. doi: 10.1186/gb-2004-5-5-223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Martienssen RA, Rabinowicz PD, O'Shaughnessy A, McCombie WR. Sequencing the maize genome. Curr Opin Plant Biol. 2004;7:102–107. doi: 10.1016/j.pbi.2004.01.010. [DOI] [PubMed] [Google Scholar]
- 66.Chandler VL, Brendel V. The Maize Genome Sequencing Project. Plant Physiol. 2002;130:1594–1597. doi: 10.1104/pp.015594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Schwartz DC, Samad A. Optical mapping approaches to molecular genomics. Curr Opin Biotechnol. 1997;8:70–74. doi: 10.1016/s0958-1669(97)80160-7. [DOI] [PubMed] [Google Scholar]
- 68.Aston C, Hiort C, Schwartz DC. Optical mapping: an approach for fine mapping. Methods Enzymol. 1999;303:55–73. doi: 10.1016/s0076-6879(99)03006-2. [DOI] [PubMed] [Google Scholar]
- 69.Aston C, Mishra B, Schwartz DC. Optical mapping and its potential for large-scale sequencing projects. Trends Biotechnol. 1999;17:297–302. doi: 10.1016/s0167-7799(99)01326-8. [DOI] [PubMed] [Google Scholar]
- 70.Lin J, Qi R, Aston C, Jing J, Anantharaman TS, et al. Whole-genome shotgun optical mapping of Deinococcus radiodurans. Science. 1999;285:1558–1562. doi: 10.1126/science.285.5433.1558. [DOI] [PubMed] [Google Scholar]
- 71.Lim A, Dimalanta ET, Potamousis KD, Yen G, Apodoca J, et al. Shotgun optical maps of the whole Escherichia coli O157:H7 genome. Genome Res. 2001;11:1584–1593. doi: 10.1101/gr.172101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhou S, Deng W, Anantharaman TS, Lim A, Dimalanta ET, et al. A whole-genome shotgun optical map of Yersinia pestis strain KIM. Appl Environ Microbiol. 2002;68:6321–6331. doi: 10.1128/AEM.68.12.6321-6331.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhou S, Kvikstad E, Kile A, Severin J, Forrest D, et al. Whole-genome shotgun optical mapping of Rhodobacter sphaeroides strain 2.4.1 and its use for whole-genome shotgun sequence assembly. Genome Res. 2003;13:2142–2151. doi: 10.1101/gr.1128803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zhou S, Kile A, Kvikstad E, Bechner M, Severin J, et al. Shotgun optical mapping of the entire Leishmania major Friedlin genome. Mol Biochem Parasitol. 2004;138:97–106. doi: 10.1016/j.molbiopara.2004.08.002. [DOI] [PubMed] [Google Scholar]
- 75.Zhou S, Bechner MC, Place M, Churas CP, Pape L, et al. Validation of rice genome sequence by optical mapping. BMC Genomics. 2007;8:278. doi: 10.1186/1471-2164-8-278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zhou S, Herschleb J, Schwartz DC. In: A single molecule system for whole genome analysis. New Methods for DNA Sequencing. Mitchelson K.R. , editor. Amsterdam: 2007. pp. 269–304. [Google Scholar]
- 77.Pruyne J, Livny M. Interfacing Condor and PVM to harness the cycles of workstation cluster. Future Generation Computer Systems. 1996;12:67–86. [Google Scholar]
- 78.Valouev A, Li L, Liu YC, Schwartz DC, Yang Y, et al. Alignment of optical maps. J Comput Biol. 2006;13:442–462. doi: 10.1089/cmb.2006.13.442. [DOI] [PubMed] [Google Scholar]
- 79.Wei F, Zhang J, Zhou S, He R, Schaeffer M, et al. The physical and genetic framework of the maize genome. PLoS Genet. 2009;5:e715. doi: 10.1371/journal.pgen.1000715. doi: 10.1371/journal.pgen.1000715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Li H, Valouev A, Schwartz DC, Waterman MS, Li LM. A quantile method for sizing optical maps. J Comput Biol. 2007;14:255–266. doi: 10.1089/cmb.2006.0006. [DOI] [PubMed] [Google Scholar]
- 81.Jing J, Lai Z, Aston C, Lin J, Carucci DJ, et al. Optical Mapping of Plasmodium falciparum Chromosome 2. Genome Res. 1999;9:175–181. [PMC free article] [PubMed] [Google Scholar]
- 82.Gardner MJ, Tettelin H, Carucci DJ, Cummings LM, Aravind L, et al. Chromosome 2 sequence of the human malaria parasite Plasmodium falciparum. Science. 1998;282:1126–1132. doi: 10.1126/science.282.5391.1126. [DOI] [PubMed] [Google Scholar]
- 83.Riera-Lizarazu O, Vales MI, Kianian SF. Radiation hybrid (RH) and HAPPY mapping in plants. Cytogenet Genome Res. 2008;120:233–240. doi: 10.1159/000121072. [DOI] [PubMed] [Google Scholar]
- 84.Okagaki RJ, Jacobs MS, Stec AO, Kynast RG, Buescher E, et al. Maize centromere mapping: a comparison of physical and genetic strategies. J Hered. 2008;99:85–93. doi: 10.1093/jhered/esm111. [DOI] [PubMed] [Google Scholar]
- 85.Anderson LK, Salameh N, Bass HW, Harper LC, Cande WZ, et al. Integrating genetic linkage maps with pachytene chromosome structure in maize. Genetics. 2004;166:1923–1933. doi: 10.1534/genetics.166.4.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Koumbaris GL, Bass HW. A new single-locus cytogenetic mapping system for maize (Zea mays L.): overcoming FISH detection limits with marker-selected sorghum (S. propinquum L.) BAC clones. Plant J. 2003;35:647–659. doi: 10.1046/j.1365-313x.2003.01829.x. [DOI] [PubMed] [Google Scholar]
- 87.Riera-Lizarazu O, Vales MI, Ananiev EV, Rines HW, Phillips RL. Production and characterization of maize chromosome 9 radiation hybrids derived from an oat-maize addition line. Genetics. 2000;156:327–339. doi: 10.1093/genetics/156.1.327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Page BT, Wanous MK, Birchler JA. Characterization of a maize chromosome 4 centromeric sequence: evidence for an evolutionary relationship with the B chromosome centromere. Genetics. 2001;159:291–302. doi: 10.1093/genetics/159.1.291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Imelfort M, Duran C, Batley J, Edwards D. Discovering genetic polymorphisms in next-generation sequencing data. Plant Biotechnol J. 2009;7:312–317. doi: 10.1111/j.1467-7652.2009.00406.x. [DOI] [PubMed] [Google Scholar]
- 90.Huang X, Feng Q, Qian Q, Zhao Q, Wang L, et al. High-throughput genotyping by whole-genome resequencing. Genome Res. 2009;19:1068–1076. doi: 10.1101/gr.089516.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Nagarajan N, Read TD, Pop M. Scaffolding and validation of bacterial genome assemblies using optical restriction maps. Bioinformatics. 2008;24:1229–1235. doi: 10.1093/bioinformatics/btn102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Dimalanta ET, Lim A, Runnheim R, Lamers C, Churas C, et al. A microfluidic system for large DNA molecule arrays. Anal Chem. 2004;76:5293–5301. doi: 10.1021/ac0496401. [DOI] [PubMed] [Google Scholar]
- 93.Anantharaman TS, Mishra B, Schwartz DC. Genomics via optical mapping III: contiging genomic DNA and variations. 1998. Courant Technical Report. [PubMed]
- 94.Anantharaman T, Mishra B, Schwartz D. Genomics via optical mapping. III: Contiging genomic DNA. Proc Int Conf Intell Syst Mol Biol. 1999:18–27. [PubMed] [Google Scholar]
- 95.Valouev A, Schwartz DC, Zhou S, Waterman MS. An algorithm for assembly of ordered restriction maps from single DNA molecules. Proc Natl Acad Sci U S A. 2006;103:15770–15775. doi: 10.1073/pnas.0604040103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Valouev A, Zhang Y, Schwartz DC, Waterman MS. Refinement of optical map assemblies (original paper). Bioinformatics. 2006:1217–1224. doi: 10.1093/bioinformatics/btl063. [DOI] [PubMed] [Google Scholar]
- 97.Huang X, Miller W. A time-efficient, linear space local similarity algorithm. Adv Appl Math. 1991;12:337–357. [Google Scholar]
- 98.Waterman M, Vingro M. Sequence comparison significance and poisson approximation. Statistical Science. 1994;9:401–418. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Using optical contigs to estimate sizes of gaps between adjacent FPC contigs. (A) Restriction map view (box = restriction fragment) of FPC contigs aligned to the AGP Chr 6 pseudomolecule; the gap is highlighted. (B) Cartoon showing the alignments of FPC contigs ct280 and ctg281 to Chr6 pseudomolecule. A gap of unknown size is illustrated, with green lines showing alignment to the pseudomolecule. Dashed lines show alignments of ct280 and ctg281 against OMcontig_6 (gold track) and a large gap. (C) Restriction map view of OMcontig_6, locating the large gap within the grayed restriction fragments. FPC gap sizes are calculated as: Gap (kb) = [|(right optical coordinate)−(left optical coordinate)|−|(right sequence coordinate)−(left sequence coordinate)|]/1000.
(0.30 MB PDF)
Ordering FPC contig sequence pseudomolecules based on the map alignments between optical maps and the in silico maps of the FPC contig sequence pseudomolecules.
(0.10 MB PDF)
Discordances in the well-aligned map segments between optical maps and the in silico maps of the B73 RefGen_v1 reference chromosomes.
(0.25 MB PDF)
FPC gap estimations based on map alignments between optical maps and the in silico maps of FPC contig sequence pseudomolecules and B73 RefGen_v1 reference chromosomes.
(0.12 MB PDF)