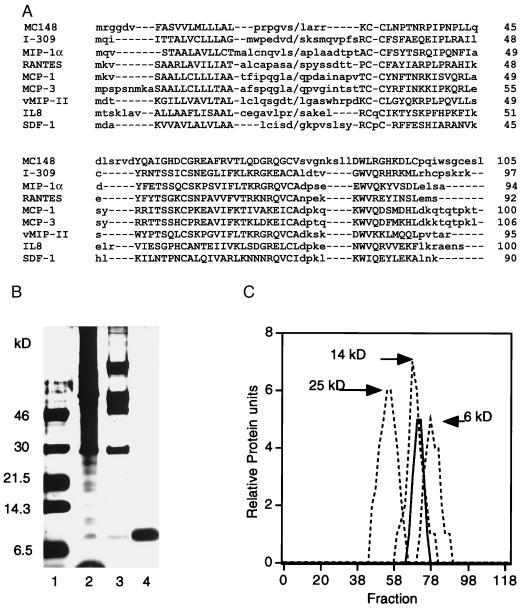
Figure 1.
Primary structure comparisons and purification of MC148P. (A) The alignment of the deduced amino acid sequence of MC148P with those of CC (I-309, MIP1-α, RANTES, MCP-1, MCP-3) and CXC (IL8, SDF-1) chemokines and HHV 8 vMIP-II chemokine homolog was performed with the macaw multiple alignment program. Upper- and lowercase letters refer to conserved and nonconserved amino acids, respectively; dashes are alignment gaps; back slashes are signal peptide cleavage sites; and numbers on the right are amino acid numbers. The gap in the NH2-terminal region of MC148 may have special significance with regard to the absence of agonist activity. (B) Purification. SDS/PAGE and silver staining of unreduced MC148P at successive purification steps. Lanes: 1, markers; 2, clarified supernatant; 3, pooled fractions from heparin column; 4, pooled fractions from SP column. (C) Elution from a G50 Sephadex column of MC148P (solid line) and the following markers (dashed lines): chymotrypsinogen, 25 kDa; ribonuclease A, 14 kDa; insulin, 6 kDa.