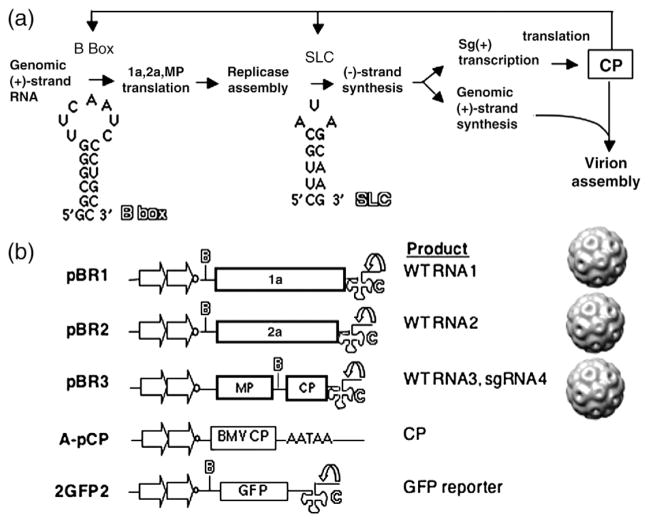
Fig. 1.
Model for the study and schematics of key constructs. (a) A model for the interaction between the BMV CP and various RNA elements in the BMV RNAs. The 5′-UTRs of BMV RNA1 and RNA2 contain a conserved motif named the B box, and the 3′-UTRs contain the SLC motif. (b) Constructs used in this study. Plasmids pBR1, pBR2, and pBR3 result in the transcription of the three BMV RNAs, which are individually encapsidated into separate virions. The plasmids are engineered to have tandem copies of the cauliflower mosaic virus 35S promoter (straight arrow) and a cis-acting ribozyme sequence (curved arrow) that will generate the authentic 3′ end of the BMV RNAs. The locations of the RNA motifs of the B box and SLC are identified by the letters B and C in outline fonts. Plasmid pCP was used for the transient expression of BMV CP from tandem copies of the 35S promoter and a translational enhancer 5′ of the BMV capsid coding sequence. The 3′-untranslated sequence contains a polyA processing signal (AATAA). Reporter RNA 2GFP2 contains the GFP coding sequence flanked by the 5′-UTR and 3′-UTR of BMV genomic RNA2.