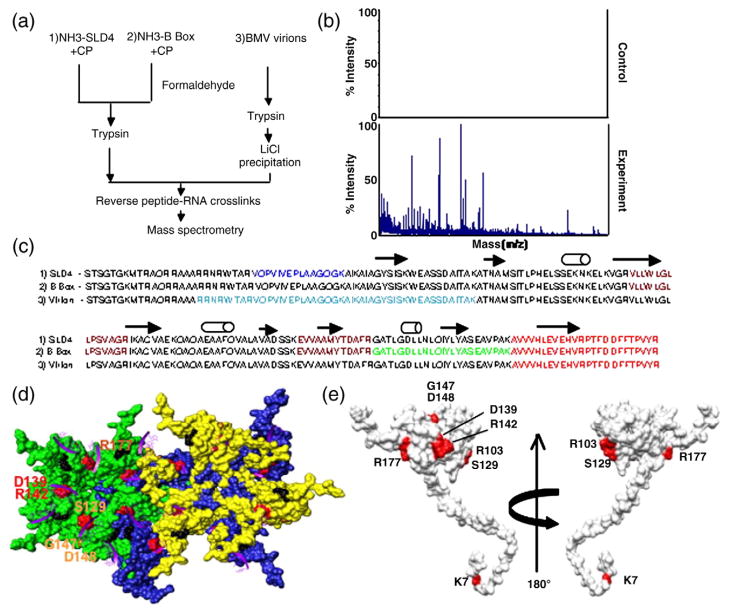
Fig. 2.
Mapping of the RNA-binding residues in the BMV CP. (a) Schematic of the RCAP protocol used to map the CP peptide–RNA interaction. Three RNA samples were analyzed as diagrammed. (b) Representative MALDI-ToF MS spectra showing the signals obtained from a control sample and a sample of the CP cross-linked to the SLD4 RNA. Both were treated identically except that the control was not subjected to formaldehyde. Signals that are three standard errors above the background were considered to be significant. (c) Sequence of the BMV CP from residues 2–189 with the peptides identified by the three RNAs in color. The peptide cross-linked to all three RNAs is shown in red. The peptide that was cross-linked to the B box and SLD4 is shown in maroon. The N-terminal methionine was not included because it was processed from the mature capsid.18 (d) Modeling of CP interaction with virion RNA. The region in the globular domain of the CP is emphasized since it was preferentially used to interact with SLD4 and the B box. The subunits that form the pentamer and hexamer are shown in green, blue, and yellow to allow better visualization of the subunits. The locations of several of the potential RNA-binding residues are identified by the names of the residues. The purple ribbons represent the fragments of RNAs seen in the CCMV crystal structure that are positioned onto the structure of the BMV capsid. The amino acids that could contact viral RNA are shown in the context of a hexamer of the BMV capsid. (e) A view of the residues in a monomer of the BMV CP that were subjected to mutational analysis.