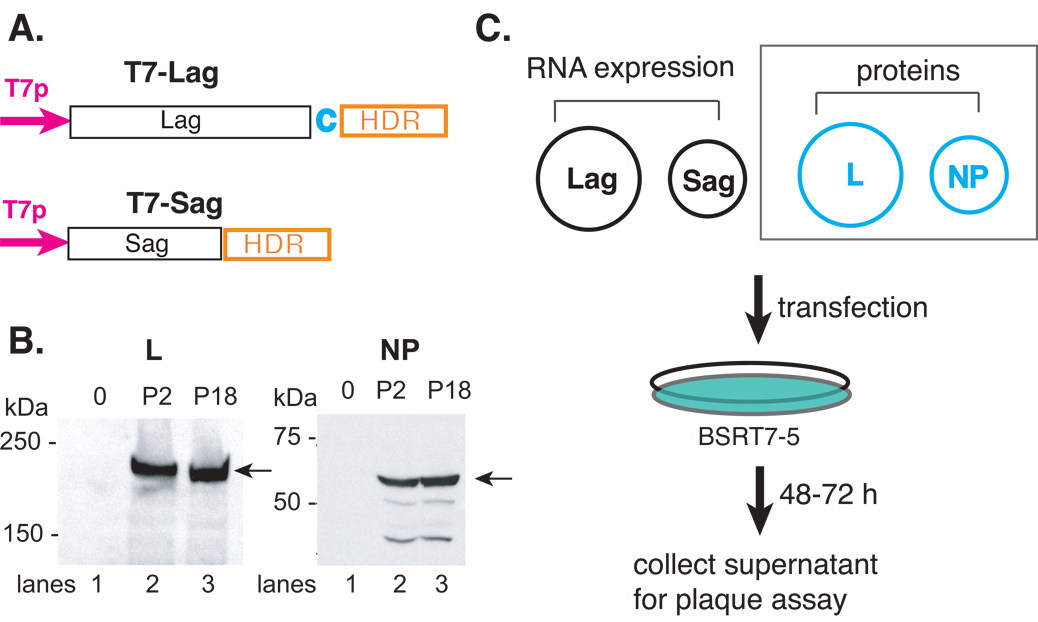
Fig. 2.
Schematic illustration of the reverse genetics systems to generate infectious rP2 and rP18 viruses from plasmid DNA transfection. (A) Schematic illustration of the RNA expression vectors of the full-length antigenomic strands of the S (pT7-Sag) and L (pT7-LagC) segments. The viral RNA segment was cloned in the antigenomic orientation immediately downstream of the T7 promoter (T7p) and upstream of the hepatitis delta ribozyme (HDR) and T7 terminator (T7t) sequences. A nonviral cytosine (in red) was added after the viral L sequence in order to enhance the self-cleavage activity. (B) The expression of the L and NP proteins. The 293T cells were transfected with an empty vector (lane 1) or vector encoding the L (left panel) or NP (right panel) gene of either the P2 (lane 2) or P18 (lane 3) virus. Western blot analyses was conducted using rabbit sera raised against specific short peptides within the L or NP protein. Peptide sequences are available upon request. (C) Schematic illustration of the reverse genetics systems. Two RNA expression plasmids (pT7-Sag and pT7-LagC) and two protein-expression plasmids of the NP and L proteins were used to transfect BSRT7-5 cells, which constitutively express the T7 polymerase protein. Between 48 and 72 hpi, supernatants were collected and plaque assay conducted on Vero cells. The two protein-expression plasmids (NP and L) (shown in dashed square) can be omitted from the reconstitution without compromising the amount of virus production.