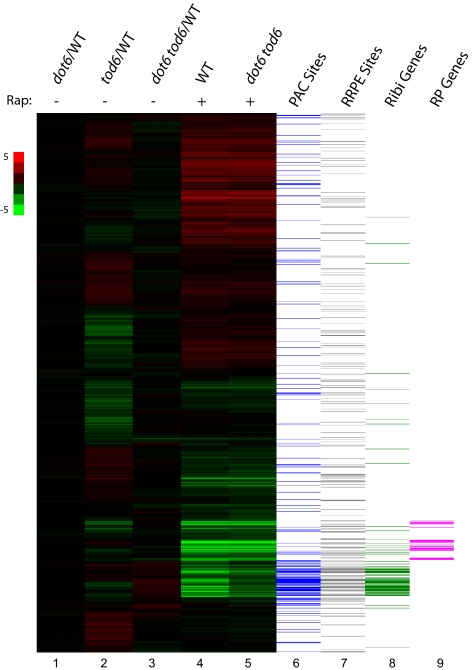
Fig. 1.
Dot6 and Tod6 specifically affect repression of ribosome biogenesis genes. Lanes 1–3: relative steady-state expression of yeast genes in mutant versus WT cells grown in rich medium. RNA isolated from Y3705 (dot6), Y3706 (tod6), or Y3707 (dot6 tod6) grown to early log phase in SC plus 2% glucose medium was labeled with Cy3 and mixed with Cy5-labeled RNA from Y2092 (WT) similarly grown and hybridized to Agilent microarray chips. The heat map organized by unsupervised hierarchical clustering shows the relative expression of each of 5,600 yeast genes (horizontal lines) in mutant versus WT (scale shown on the left). Lanes 4 and 5: strains Y2092 (WT) and Y3707 (dot6 tod6) were grown in SC plus 2% glucose medium to a density of OD600 = 0.25, at which point rapamycin was added to a final concentration of 100 nM. Heat map represents expression levels of yeast genes 80 min after rapamycin addition relative to cells just before addition for each of the two indicated strains. Lanes 6 and7 denote the presence of PAC (GCGATGAGNT) or RRPE (TGAAAATTT) motifs within 500 bp upstream of the transcriptional start site of the corresponding yeast gene, and lanes 8 and 9 indicate whether the corresponding genes are annotated as ribosome biogenesis (Ribi) or ribosomal protein (RP) genes.