Figure 2.
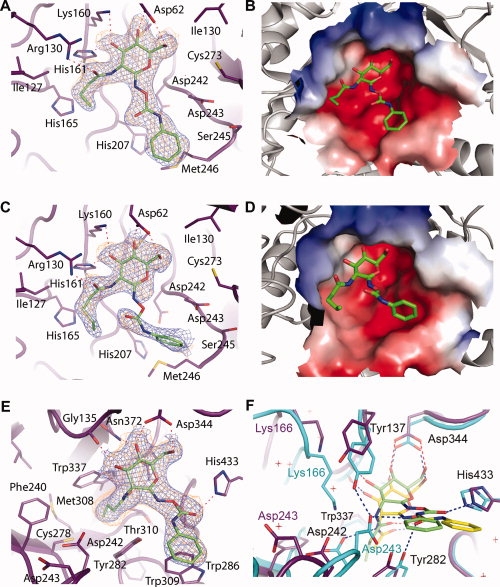
Crystal structures of N-butyryl-PUGNAc and N-valeryl-PUGNAc bound to VcNagZΔSC, and N-butyryl-PUGNAc bound to BtGH84. The active site of VcNagZΔSC binds N-butyryl-PUGNAc (A,B) and N-valeryl-PUGNAc (C,D) primarily through hydrogen bonding (dashed lines) with the pyranose ring of these inhibitors. The N-phenylcarbamate moiety of each inhibitor does not form direct hydrogen bonds with the enzyme and appear mobile. The average B-factor for the pyronose ring of N-butyryl-PUGNAc is 28 Å2, whereas its phenyl ring has an average value of 37 Å2. The pyronase ring of N-valeryl-PUGNAc has an average B-factor of 36.3 Å2; however, the average value for the phenyl ring was 63 Å2. Because of the high average B-factor and lack of continuous electron density, atoms of the N-phenylcarbamate of N-valeryl-PUGNAc are not included in PDB entry 3gsm; however, a refined position is shown here. Electrostatic surface potential maps (negative, red; positive, blue) calculated using APBS30 show the N-butyryl and N-valeryl extensions bound within a large, solvent accessible binding groove within the active site. E: BtGH84 bound to N-butyryl-PUGNAc. The inhibitor is bound via hydrogen-bonds (dashed lines), including one between the carbonyl oxygen of the N-phenylcarbamate extension and His433, which appears to order this moiety relative to the flexible binding observed within VcNagZΔSC. F: Superposition of N-butyryl-PUGNAc and PUGNAc bound31 complexes of BtGH84 reveals large structural distortions and displacement of N-butyryl-PUGNAc within the active site. Blue electron densities are maximum-likelihood weighted 2Fobs − Fcalc syntheses contoured at 0.24 e/Å3. Orange densities are maximum-likelihood weighted Fobs − Fcalc syntheses contoured at 0.11 e/Å3 following refinement with inhibitor models omitted. Superposition was carried out using CCP4.32 Figures were created with PyMOL.33
