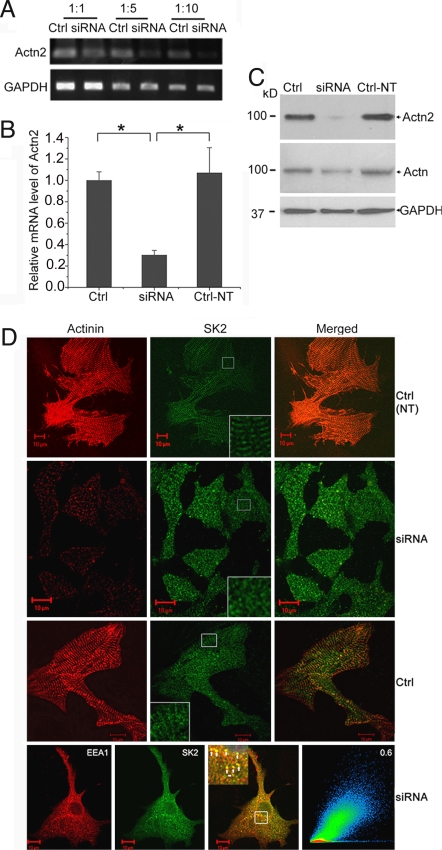
Fig. 4.
α-Actinin2 cytoskeletal protein is required for normal SK2 channels localization in neonatal cardiomyocytes. (A and B) mRNA level of α-actinin2 was analyzed by semiquantitative RT-PCR using different dilutions of cDNA (A) and real-time RT-PCR (B) in neonatal cardiomyocytes treated with specific siRNA targeted against α-actinin2 or scrambled siRNA (Ctrl) and nontransfected cells (Ctrl-NT). There was a significant reduction in α-actinin2 mRNA level by specific siRNA compared with scrambled siRNA according to semiquantitative RT-PCR (A) as well as real-time RT-PCR (B). GAPDH mRNA level was used as control. Values are means ± SEM (n = 6; *, P < 0.05). (C) Western blot analysis showing α-actinin2 protein level detected by monoclonal rabbit anti-actinin2 antibody (Top) and monoclonal anti–α-actinin (sarcomeric) antibody (Middle) and GAPDH (Bottom) for loading control in cultured neonatal mouse cardiomyocytes with different treatments. Consequently, α-actinin2 protein level detected using rabbit monoclonal anti–α-actinin2 antibody was reduced significantly after transfection by specific siRNA. (D) Confocal microscopic images of immunostaining using anti-SK2 and anti–α-actinin antibodies for siRNA-treated or control groups. Cardiomyocytes transduced by siRNA specific to α-actinin2 show a significant decrease in the α-actinin2 staining; the SK2 channels were expressed but displayed abnormal pattern with localization mainly in the cytosol. Insets: Higher magnifications of the original marked area. Bottom row shows a high correlation between the abnormally localized SK2 channels with EEA1 in cardiomyocytes treated with siRNA.