Figure 2.
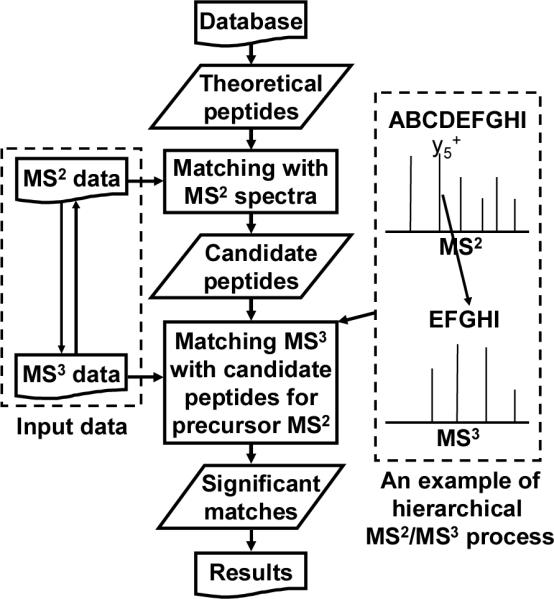
Diagram of hierarchical MS2/MS3 database search algorithm in MassMatrix. Theoretical peptides are created from the protein database by in silico digestion and modification with specified PTMs. MassMatrix first matches experimental MS2 spectra to the theoretical peptides to obtain candidate peptide matches. MassMatrix then matches the corresponding MS3 spectra against each candidate peptide matches from the MS2 search results. In this manner, the MS2 and MS3 spectral data are used in concert to obtain the peptide matches.
