Figure 2. Potential Vps60 binding sites on Vta1NTD.
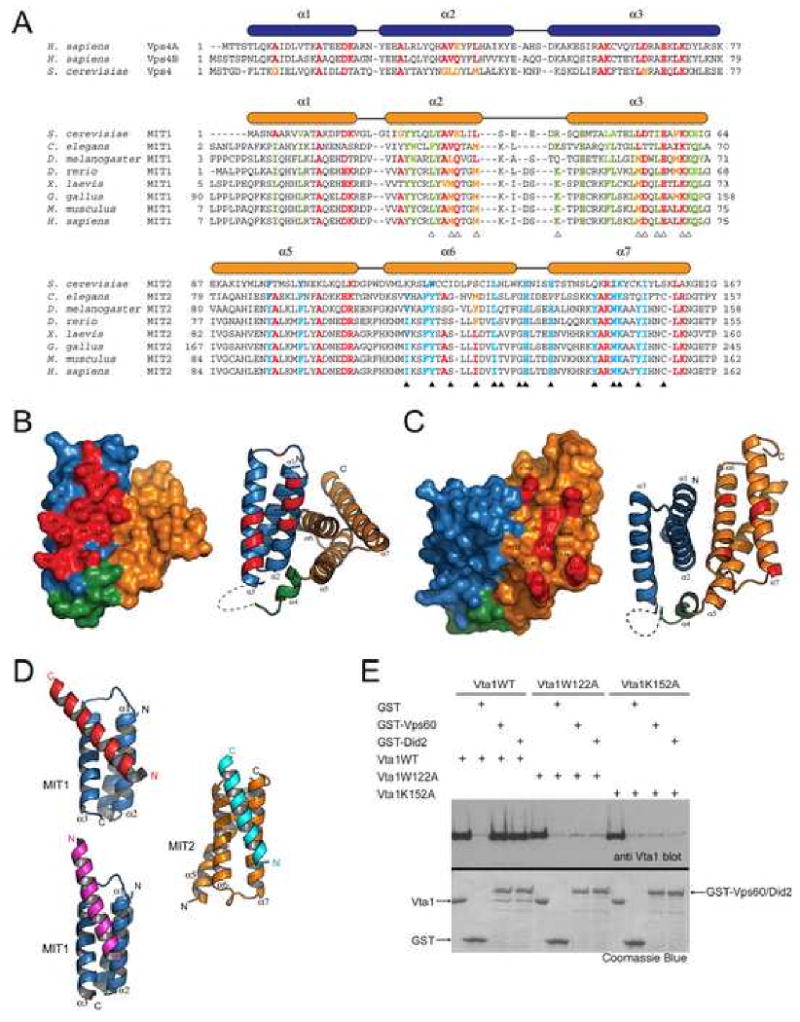
(A) Structure-based sequence alignment for the MIT domains in Vps4 and the two MIT motifs in Vta1. Secondary structure elements are shown above the sequences. Residues conserved in all MIT domains are colored red and yellow. Residues conserved only in MIT1 are colored green; and residues conserved only in MIT2 are colored blue. Residues involved in the potential functional surfaces of MIT1 and MIT2 are marked by white and black triangles, respectively. (B, C) Conserved molecular surfaces on MIT1 (B) and MIT2 (C). Surface and ribbon representations of molecules in the same orientation are shown on the left and right panels, respectively. MIT1 is colored blue; helix α4 is colored green; and MIT2 is colored orange. Conserved molecular surfaces are colored red and underlying residues are labeled. (D) The two conserved surfaces are involved in interacting with an α-helix from molecules in the crystal lattice. MIT1 is colored blue and MIT2 is colored orange. The helices they interact with in the crystal lattice are colored red, magenta, and cyan, respectively. (E) Residues in MIT2 are important for Vps60 and Did2 binding. GST or GST-tagged Vps60/Did2 was used to pull down purified wild-type Vta1 or mutants as indicated. Proteins retained on the beads were analyzed by SDS-PAGE and visualized by Coomassie blue staining or western blotting with anti-Vta1 antibody (see the accompanying study (Azmi et al., 2007) for anti-Vta1 polyclonal antibody production). Figures 2B, 2C and 2D are prepared with Pymol (DeLano Scientific LLC).
