Fig. 5.
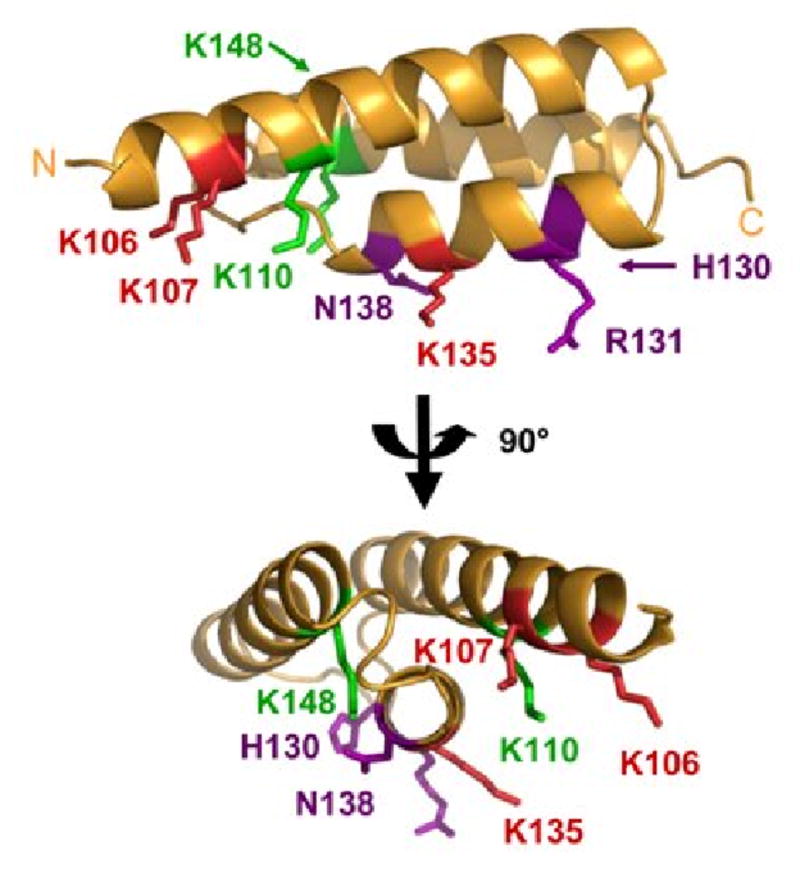
Solution contacts of Efb(-C) and C3d mapped onto the Efb-C crystal structure. The results of differential lysine acetylation studies for free and bound Efb(-C) are superimposed on the Efb-C crystal structure (orange ribbon); two orthogonal views are shown and were generated by rotating in the plane of the page. Residues K106, K110, H130, R131, K135, N138 and K148 of Efb-C were previously identified by x-ray crystallography as contributing to formation of Efb-C/C3d. K110 was solvent accessible in fragment QKLIQAQNLVRE (109-119), and K148 was fully acetylated in the fragment KVKKMVLQERIDNVL (145-159), both of which were derived from the Efb-C/C3d complex. K110 and K148 are therefore shown in green since they do not appear to interact strongly with C3d. Oppositely, K106 was protected in fragment NKPAAKTDATIKKEQKLIQAQNL (95-117) and K135 was protected in the fragment FEKTHTVSAHRKAQKAVNLVSFE (121-143) obtained from the complex. Thus, the five lysine residues that are protected in the protein complex are (shown in red): K96, K100, K106, K107 and K135. Since K96 and K100 lie within a disordered region of the Efb-C structure, these residues cannot be displayed. H130, R131, and N138 are shown in purple since these sidechains do not react with NHS-Ac.
