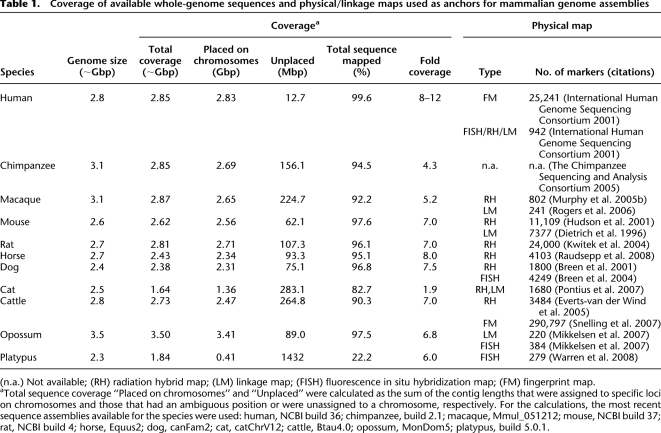
Table 1.
Coverage of available whole-genome sequences and physical/linkage maps used as anchors for mammalian genome assemblies
(n.a.) Not available; (RH) radiation hybrid map; (LM) linkage map; (FISH) fluorescence in situ hybridization map; (FM) fingerprint map.
aTotal sequence coverage “Placed on chromosomes” and “Unplaced” were calculated as the sum of the contig lengths that were assigned to specific loci on chromosomes and those that had an ambiguous position or were unassigned to a chromosome, respectively. For the calculations, the most recent sequence assemblies available for the species were used: human, NCBI build 36; chimpanzee, build 2.1; macaque, Mmul_051212; mouse, NCBI build 37; rat, NCBI build 4; horse, Equus2; dog, canFam2; cat, catChrV12; cattle, Btau4.0; opossum, MonDom5; platypus, build 5.0.1.