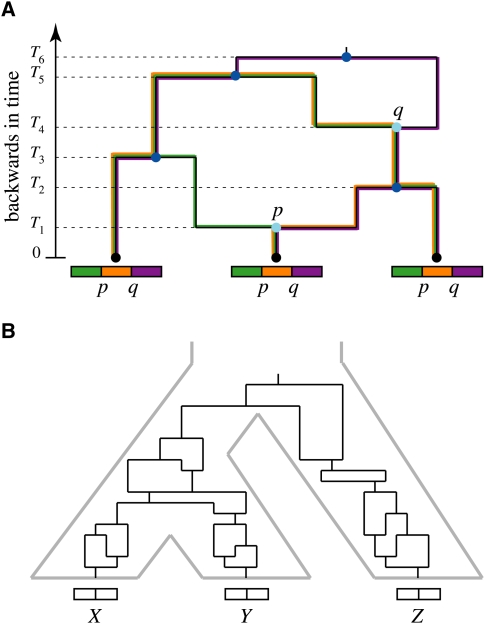
Figure 4.
(A) An ancestral recombination graph (ARG) for three individuals. As the graph is followed backward in time, edges can be seen to merge (times T2, T3, T5, and T6, dark blue), where descendant lineages coalesce at common ancestors, or to split (times T1 and T4, light blue), where descendants derive from recombining ancestors. (By assumption, at most one event occurs at each instant in time, so all internal nodes have three adjacent edges.) Ultimately, coalescences overwhelm recombinations and the graph is reduced to a single node (the global most recent common ancestor). Each recombination node is associated with a point along the sequence at which the recombination event occurred (p or q). For any nonrecombining segment of the sequence (green, orange, and purple), a genealogy can be extracted by choosing the left or right edge exiting each recombination node, depending on the position of the segment relative to the recombination point. Thus, the graph defines a set of marginal genealogies for the nonrecombining segments (traced here in colors matching the segments). (B) A “phylogenetic ARG” for three individuals from different species (X, Y, and Z). This graph is the same as the ARG for individuals in a single interbreeding population except that both recombination and coalescence events are prohibited from occurring across species boundaries (gray). Its marginal genealogies will in general exhibit differences in branch length and topology that reflect ancestral population dynamics and historical recombination.