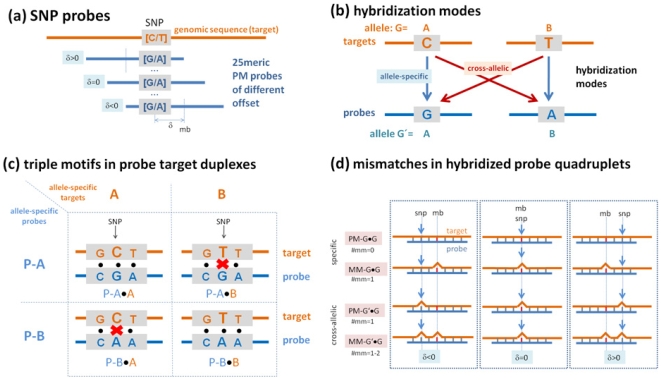
Figure 1. Probe design and hybridization modes for SNP detection.
(a) Each SNP (for example [C/A]) is probed by 25meric probes of complementary sequence. Different offsets δ of the SNP position relative to the middle base (mb) of the probe sequence are used. In addition, each PM probe is paired with one MM probe the middle base of which mismatches the target sequence (not shown). (b) The allele-specific probes intend to detect the respective targets via allele-specific binding which however competes with cross-allelic hybridization of targets of the alternative allele (see also the reaction equation Eq. (6)). (c) Both hybridization modes give rise to four different types of probe/target duplexes formed by the two allele-specific probes. The figure shows the respective base pairings for a selected SNP-triple which consists of the SNP [C/T] and its nearest neighbors. Mismatched non-canonical pairings are indicated by crosses. (d) Each box includes one probe-quartet which consists of two PM/MM-probe pairs interrogating either targets of allele G = A or targets of allele G' = B and vice versa (i.e. G = B and G' = A). Only targets of one allele are assumed to be present as in the sample. They hybridize to the probes of both allele sets forming either specific or cross-allelic duplexes, respectively. The three selected probe quartets differ in the offset δ of the SNP position (see arrows and part a of the figure) relatively to the middle base of the probe. The different combinations give rise to different numbers and positions of mismatched pairings which are indicated by the bulges. Their number varies between #mm = 0 and #mm = 2 in dependence on the probe type, hybridization mode and offset position. Complete probe-sets use 10 probe quartets.