Figure 5.
Specific Binding of Nuclear Proteins from Human Cells to DNA Sequences in the Boundary Region
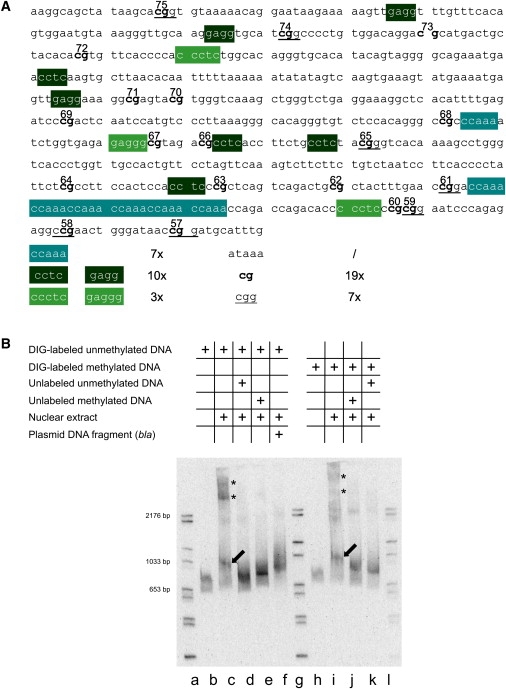
(A) Nucleotide sequence in the transition zone between fully methylated and unmethylated DNA in the FMR1 5′-upstream region. Several DNA motifs and a 5′-(CCAAA)6-3′ repeat are indicated.
(B) Nuclear extracts from human HCT116 cells (2.64 μg of protein) were incubated with the DIG-labeled 630 bp DNA fragment from the boundary region in the FMR1 5′-upstream genome segment as indicated. The reaction products were analyzed by electrophoretic mobility shift assays. Lanes a, g, and l: Marker DNA lanes with DIG-labeled DNA fragments of sizes as marked. Lane b: DIG-labeled 630 bp fragment incubated without the addition of nuclear extracts; lane c shows the same after incubation with nuclear extracts. Lane d: conditions as in lane c, except that a > 2000-fold excess of an unlabeled 630 bp fragment was added as specific competitor. Lane e: conditions as in lane c, except that a > 2000-fold excess of an unlabeled M.Sss I-premethylated 630 bp DNA fragment from the transition region was used for competition. Lane f: conditions as in lane d, except that the competitor was a 595 bp DraI-RsaI fragment from the bla gene of E. coli plasmid pcDNA3.1 (+). In lanes h–k, an M.Sss I-premethylated 630 bp DNA fragment from the transition region was used for binding or competition experiments. Competition experiments were performed as indicated in the graph. Electrophoretic mobility was toward the bottom. After electrophoresis in a precast 6% polyacrylamide gel, the DNA bands were visualized as described in Material and Methods.
In (B), the presumably unspecific complexes are designated by asterisks (∗), the specific ones by arrows.
