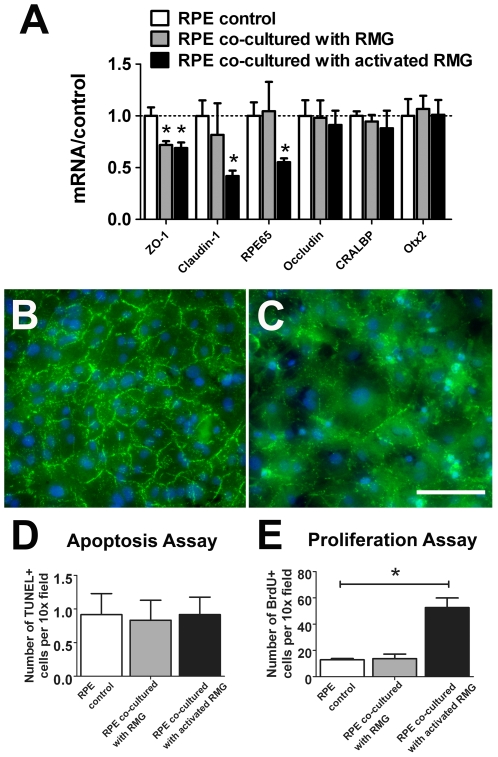
Figure 2. Primary RPE cells are altered in gene expression, structure and proliferation following co-culture with retinal microglia (RMG).
(A) Quantitative RT-PCR comparing mRNA levels of visual cycle (RPE65, CRALBP) and junctional proteins (ZO-1, Claudin-1, Occludin) in RPE cells cultured alone (control, white bars), or co-cultured with either unactivated RMG (gray bars) or RMG pre-activated with LPS (black bars). Results were normalized relative to levels in unexposed RPE cells. (B–C) ZO-1 labeling (green) in confluent primary RPE cells co-cultured in the absence (B) and the presence of activated RMG (C). Co-culture with RMG induced the loss of discrete localization of ZO-1 in RPE tight junctions. Scale bar = 50 µm. (D) RPE cell apoptosis, as measured by TUNEL labeling, in the presence of conditioned media from unexposed RPE cells (white), or from RPE cells co-cultured with unactivated (gray), or activated RMG (black) (*p = <0.0001, n = 8 replicates). (E) RPE cell proliferation, as measured by BrdU staining, in the presence of unexposed RPE cells (white) or from RPE cells co-cultured with unactivated RMG (gray), or activated RMG (black). (p = 0.973, n = 12 replicates for each condition.) Significant differences (p<0.05) are indicated with *.