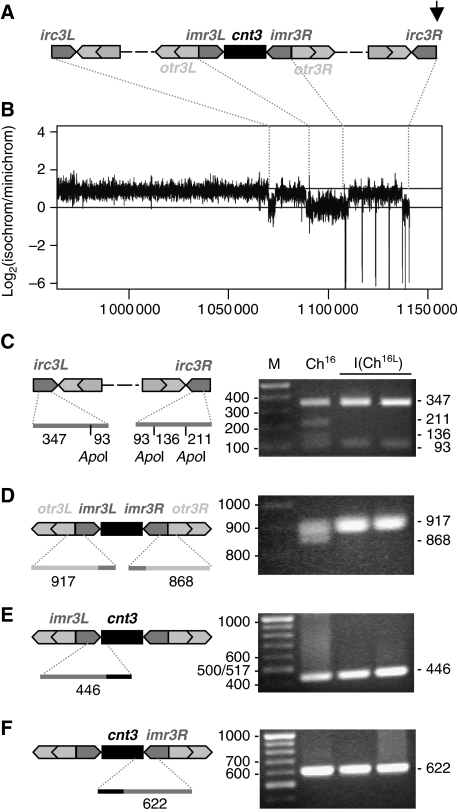
Figure 5.
Analysis of the isochromosome centromere. (A) Schematic of predicted centromere structure of Cen-Ch16 based on homologous cen3, with the break point of Ch16-RMGAH and Ch16-YAMGH-derived LOH colonies, indicated by an arrow. cnt3, innermost (imr3), outer (otr3) and irc3 repeats are shown. (B) Log2 ratio of the fold coverage from Illumina GAII sequencing across isolated isochromosome and minichromosome DNA (log2 of isochromosome/minichromosome) from strains TH4313 and TH2125, respectively. Sequence data aligned to S. pombe ChIII reference sequence at positions indicated (C) PCR amplification of irc3L and irc3R using irc3-R and irc3-F primers, digested using ApoI, (D) PCR amplification of imr3–otr3 junctions using imr-out and dh primers (E) PCR amplification of imr3L–cnt3 junction using primers cnt3-L and imr3-in (F) PCR amplification of cnt3–imr3R junction using primers cnt3-R and imr3-in, separated on a 2% agarose gel and stained with EtBr. Diagnostic PCR product sizes are shown. PCR amplification was carried out as described previously (Nakamura et al, 2008).