Abstract
Influenza infection remains a leading cause of infectious disease-mediated morbidity and mortality. Accumulating evidence indicates that most variants of seasonal and pandemic influenza have developed resistance to conventional therapies. Such information has spawned new interest in identifying novel approaches to target influenza. Our laboratories have been developing a new strategy of Host-Oriented Therapeutics, which seeks to target host molecules in a safe and effective manner that prevents the virus from causing disease. Using an improved discovery technology, Random Homozygous Gene Perturbation (RHGP), we identified the PTCH1 protein as an essential host target that critically controls influenza virus infection. We further demonstrated that targeted intervention against PTCH1 using antibodies or siRNA decreases influenza infection. Finally, we demonstrated the involvement of PTCH1 in influenza infection outside of the laboratory by showing that genetic variations of PTCH1 relate to decreased disease morbidity in the field. Altogether, these findings have important implications for the development of novel, host-directed therapeutics to improve influenza disease management.
Keywords: Influenza virus, random homozygous gene perturbation (RHGP), PTCH1, resistance to therapies, siRNA
Introduction
Influenza remains one of largest causes of infectious disease-mediated suffering and death. In an average year, influenza claims 20–30,000 lives and billions of dollars in economic damages [1]. Despite its widespread prevalence, vaccination remains sporadic, and recent studies indicate vaccines may be effective in less than half of those treated [2]. Compounding this, only a handful of therapeutics have been developed to treat influenza infection and increasing evidence indicates many seasonal and pandemic (avian) strains have become resistant to these compounds, thus obviating their application [3, 4]. The inevitable outgrowth of drug-resistant strains is not unique to influenza, and the emergence of resistant strains of influenza, HIV, herpes and other viruses has forced the medical community to reconsider strategies for antiviral therapy.
Most conventional antivirals target virus-encoded pathways. The fundamental rationale has assumed that targeting viral pathways will minimize toxicity to normal host cells. However, many of the resulting compounds are indeed quite toxic and worse still, a focus on viral targets places selective pressure on the pathogen to derive variants that are resistant to therapy. This need for alternatives has driven the approach practiced herein, which seeks to develop new therapeutics that are insensitive to drug resistance and which can be applied to a broad spectrum of different virus types [5–7].
Our laboratory has been developing a concept of Host Oriented Therapeutics for Infectious Disease [8]. This approach seeks to identify host-derived targets that are mis-expressed or functionally altered in virus-infected cells. In doing so, we have postulated that the altered expression or function would provide opportunities for novel therapeutics that are safe and effective in treating viral diseases. By identifying, and then targeting, pathways that are essential to different types of influenza, we might develop a much-needed antiviral with a broad-spectrum of application. A clear gap in knowledge is the identification of safe and effective host targets and, ideally the means to prosecute those targets. To that end, we successfully utilized our core technology, Random Homozygous Gene Perturbation (RHGP). RHGP utilizes a novel gene search vector (GSV), which was designed to interrogate the entire genome and identify targets that allow host cells to resist or survive infection with influenza virus [8]. Our experimental strategy centered upon integration of the GSV at a single site in the genome, where it can either cause overexpression or loss of expression of the target gene. As such, RHGP allows us to interrogate the entire cell genome to identify different types of events that allow host cells to resist or survive influenza infection. In our present report, we utilize RHGP to identify PTCH1 as an essential host target in influenza infection. We further demonstrate that targeting intervention against PTCH1 can block influenza infection. Finally, SNP (single nucleotide polymorphism) analyses demonstrate that PTCH1 genotypes relate to disease outcome during natural outbreaks of influenza in porcine populations.
Materials and methods
Cell culture
Human MDA-MB231 breast cancer cells were purchased from the American Type Culture Collection (ATCC). The Phoenix A cell line was a gift from Dr. Nolan, Stanford University and mouse N2a cell line was a gift from Dr. Xu, Rockefeller University. MDA-MB231 cells were cultured in DMEM containing 10% FBS and N2a cells were cultured in 50% DMEM and 50% Opti-MEM media containing 5% FBS.
RHGP library generation and screening
The RHGP gene search vector was applied to generate RHGP libraries in MDCK cells as detailed [8]. RHGP libraries were infected with influenza virus A/Udorn/72 at a multiplicity of infection (MOI) of 0.1 or 0.001. Two weeks after second round of infection, genomic DNA was obtained using the BIO-RAD AquaPure Genomic DNA Isolation Kit. Self-ligated genomic DNA was concentrated and electroporated into E.coli cells and selected using chloramphenicol.
Validation of host target genes with siRNA
The human duplex siRNA homologues for PTCH1 were prepared as recommended by the manufacturer. The siRNA NP-1496 sequence (GGAUCUUAUUUCUUCGGAGUU), which targets the nucleocapsid (NP) gene of influenza virus, provided a positive control [9]. Non-targeting siRNA, siCONTROL1 provided a negative control. HEK293 cells were plated in 24-well plates at 1×105 cells per well, respectively. After 24h incubation, the cells were transfected with 20 nM of siRNA and TransIT-TKO, according to the manufacturer's instruction (Mirus). Twenty-four hours after the second round of transfection, the samples were washed with MEM followed by infection with influenza virus A/Udorn/72 (MOI 0.1). The cells were incubated for 1h with gentle rocking every 15 min. The culture medium from each well was collected 48 h post-transfection and progeny viruses in the medium were titrated using standard plaque assays.
Flow cytometry
For flow cytometric analysis, MDCK cells were suspended using trypsin and 1×105 cells were incubated on ice with 10 mg/mL antibody for 30 min. Cells were washed three times with PBS containing 1% BSA (Sigma, St. Louis, MO) in PBS on ice and incubated with FITC conjugated goat anti-rabbit (Becton Dickinson, San Jose, CA) for 30 min on ice. After washing the cells were fixed in PBS with 1% paraformaldehyde. Data were acquired using an EasyCyte Flow Cytometer (Guava Technologies, Hayward, CA) and analyzed using FlowJo analysis software.
SNPs and datasets
Seven SNP in or near the PTCH1 gene were identified and four were selected based on an acceptable allele frequency in selected pig lines and successful formatting by the external genotyping provider (Sequenom). Table 1 provides the major allele frequency of each of the selected SNPs.
Table 1.
Frequency of alleles for each of the selected SNPs in or near the PTCH gene
| SNP | Location | Major allele frequency |
|---|---|---|
| 619 | PTCH | 0.882 |
| 660 | Near PTCH | 0.966 |
| 661 | Near PTCH | 0.872 |
| 662 | Near PTCH | 0.708 |
Shown is a listing of the single nucleotide polymorphisms (SNP) within or near porcine PTCH that were used to assess robustness in influenza-infected porcine populations.
The Influenza dataset was collected in a commercial pig production system using cross-bred PIC genetics. Both tissue samples (ear notches) and phenotypic data were collected. During the study period, the farm experienced an outbreak of swine influenza virus. Four growth traits were measured: 1). Growth: As a binomial trait that compared high and low growth animals; 2) LDG: Lifetime daily gain (g/day) [(offtest wt – birth wt)/age at offtest]/1000; 3) Lifetime Daily Carcass Gain: (grams/ day); 4) Wean to offtest daily (g/day) [offtest weight – wean weight) / (offtest date – weaning date)] / 1000. The size of the effect in the presence of a single copy of the favorable allele was determined using least squares means (LSM).
Results
Identification of influenza insensitivity caused by disruption of PTCH1
To identify targets that render host cells resistant to influenza, an RHGP library was generated as described previously [8] in MDCK cells and influenza challenge performed as indicated (Figure 1A). The library was challenged by infection with influenza A/Udorn/72 to select for influenza-resistant cells. We had previously established that infection with A/Udorn/72 (MOI of 10−1) reproducibly killed all MDCK cells within 48 h (not shown). As a control, parallel cultures of mock-transduced cells were treated identically and no survivors were observed after 48 h.
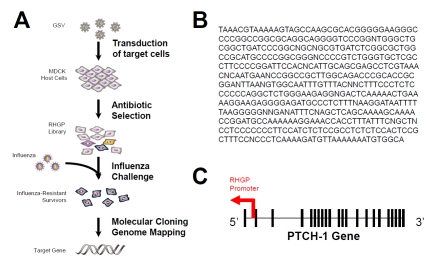
Figure 1.
Identification of PTCH1 regulation of influenza infection. (A) Shown is an overview of the experimental strategy used for RHGP-mediated identification of host targets that prevent influenza killing of MDCK cells. (B) The DNA sequence of influenza-resistant isolate R26-7 is shown. (C) The nucleotide sequence above corresponds to an antisense disruption of the canine PTCH gene.
After selecting for RHGP-transduced cells that survived multiple rounds of influenza challenge, we focused on one particular culture (R26-7), which survived influenza infection. The RHGP gene search vector allowed us to efficiently locate target genes and determine the orientation (sense or antisense) of the integration event. Genomic DNA was isolated from R26-7 cells and genomic DNA sequences flanking the RHGP vector insertion sites was sequenced and mapped against the canine genome using the UCSC Genome Browser (Figure 1B). The integration event in R26-7 occurred in canine chromosome one, in an antisense orientation, in the PTCH1 gene (Figure 1C). As such, the integration physically disrupted the first copy of PTCH1 and the resulting antisense produced by the GSV knocked down expression of the second allele.
Validation of PTCH1 using naïve cells
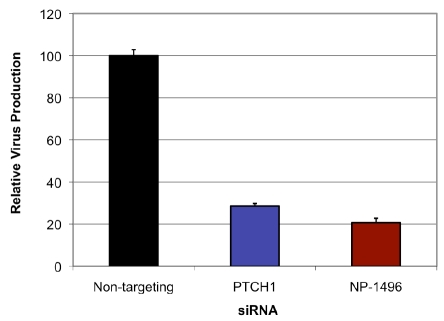
We then sought to verify PTCH1 as a candidate target that prevents influenza-mediated killing of host cells. These studies were meant to exclude artifacts that might arise from RHGP or the use of MDCK cells. Since PTCH1 had been isolated from canine-derived MDCK cells, we identified its human homologue and expressed siRNAs selective for these targets in human HEK-293 cells. Duplex siRNAs were generated and transfected into HEK293 cells (Figure 2). Non-targeting siRNAs provided a matched control for the transfection and a reference standard. NP-1496 duplex siRNAs, which target a critical influenza gene and have been demonstrated to inhibit influenza infection, provided a positive control [9]. Since influenza infection does not efficiently kill HEK293 cells, we modified our experimental protocol to measure viral titers (instead of host cell survival) as the primary endpoint for efficacy. The siRNA-treated HEK293 cells were infected with A/Udorn/72 for 48 hours and viral titers were measured by plaque assays. PTCH1 duplex siRNAs decreased influenza virus production and at levels comparable to the positive control (NP-1496 siRNAs).
Figure 2.
Validation of PTCH-1 as a host-oriented target for influenza. Confirmation of PTCH-1 involvement in the influenza life cycle was accomplished by antisense disruption of human PTCH1 in naïve human HEK293 cells. Note that siRNA targeting of PTCH1 was sufficient to inhibit influenza production by HEK293 as well as an established influenza target (NP-1496), which provide a positive control for siRNA-mediated inhibition of influenza.
Antibody targeting of PTCH1 decreases influenza infection
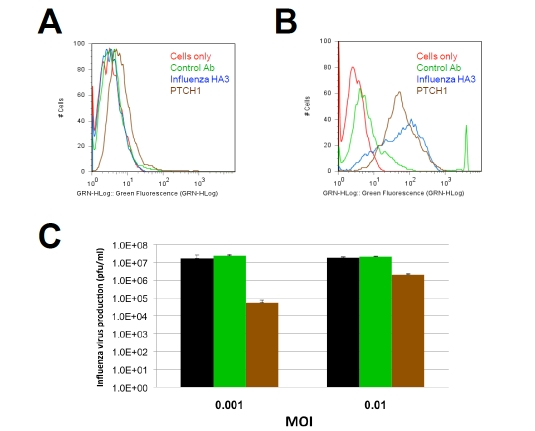
PTCH1 is a multipass transmembrane protein [10]. As such, we considered that it might provide a target for antibody-based intervention. To evaluate this possibility, affinity purified antisera were obtained and tested for their abilities to recognize PTCH1 on the surface of influenza-infected cells. MDCK cells were infected with A/Udorn influenza for 18 h prior to staining of intact cells (Figure 3). This experimental design was utilized to minimize the potential for artifacts that could arise as a result of prolonged incubation with virus or fixation, either of which could disrupt membrane integrity. Antibodies specific for viral-encoded hemagglutinin and a matched control, which does not recognize PTCH1, provided positive and negative controls, respectively. PTCH1 antibodies bound influenza-infected cells and considerably more staining was observed with infected cells (Figure 3B) than non-infected controls (Figure 3A). This outcome did not represent a loss of membrane integrity since the cells continued to exclude Trypan Blue and other vital dyes under the conditions utilized (data not shown).
Figure 3.
Antibody targeting of PTCH1 blocks influenza infection. Antibody binding to the surface of (A) control or (B) influenza-infected MDCK cells was evaluated using flow cytometry. PTCH1 staining (brown) is selectively exposed on the surface of influenza-infected cell. Matched isotype controls (green) and influenza-hemagglutinin (Blue) antibodies provided controls negative and positive controls for staining, respectively. (C) MDCK cells were infected with influenza A/Udorn/72 in the presence of absence of PTCH1 antibodies (brown bars) or isotype-matched control antibodies (green). Note that the antiviral effects of PTCH1 antibodies are more prominent at a low multiplicity of infection (MOI), suggesting PTCH1 antibodies inhibit a late stage of the viral life cycle.
Based on the abilities of PTCH1 antibodies to bind influenza-infected cells, we then asked if treatment with these antibodies might alter influenza infectivity. MDCK cells were infected with influenza in the presence or absence of PTCH1 antibodies. Culture supernatants were harvested and the number of infectious influenza particles quantified using standard plaque assays. These studies demonstrated that PTCH1 antibodies decreased influenza propagation by 2–3 logs (100–1000 fold) as compared with untreated cells or controls that had been treated with non-specific antisera (Figure 3C). We then asked if this outcome might represent antibody inhibition of initial infection or later stages of the viral life cycle. This question was addressed by repeating infection at a higher MOI of 0.1 rather than 0.001. We reasoned that if PTCH1 antibodies comparably inhibited influenza at a relatively high MOI, they might be blocking initial attachment or entry into the infected cells. On the other hand, if the antibodies impacted a later stage of the life cycle (e.g., budding or release), then inhibitory activity would be decreased at a higher MOI. Consistent with the latter idea, the antiviral effect of PTCH1 antibody treatment was decreased at a higher MOI, imparting a one log (10 fold) decrease in influenza. These results suggest that PTCH1 may provide a suitable target for antibody therapy and that the antibodies appear to be blocking a relatively late stage of the viral life cycle.
Translating PTCH1 from the laboratory to influenza infection in the field
Based on intriguing findings with cell-based assays, we asked if PTCH1 would relate to influenza susceptibility outside of the laboratory. For this, we performed an association study of PTCH SNP and robustness traits in swine populations during a swine influenza virus outbreak. Specifically, we asked if the PTCH1 SNPs were associated with changes in swine weight gain, which is an understood outcome of influenza infection in pigs.
A series of different SNP within or near PTCH1 (known as 619, 661, 662 and 660) were selected based on several criteria, including level of significance, allele frequency, additive vs. dominance effects, and size of effect (phenotypic standard deviation). We then asked if the SNPs were associated with influenza resistance in the field. For this, DNA from pigs exposed to virus infections in the field was extracted, genotyped for each SNP, and statistical associations between SNP genotype and performance traits were determined. The study utilized a dataset from a large farm that had undergone a swine influenza outbreak during the period of dataset collection. Our analyses revealed two different PTCH1 SNPs (619 and 660) with increased average daily weight gain and weight gain since time of weaning (Table 2).
Table 2.
PTCH1 polymorphisms relate to increased weight gain of porcine populations in the context of a swine influenza outbreak
| Marker | Lifetime Daily Gain (grams/day) | Wean End Daily Gain (grams/day) |
|---|---|---|
| 619 | 23.69 (0.023) | 21.88 (0.036) |
| 660 | 14.19 (0.047) | 15.16 (0.033) |
| 661 | 11.93 (0.149) | 13.53 (0.105) |
| 662 | 6.81 (0.120) | 7.81 (0.076) |
The effect of PTCH1 polymorphisms on the growth (grams/day) of one copy of favorable allele (relative to the major allele). All results are shown as the relative increases in weight gain relative to the major allele (p value). Lifetime Daily Gain = Final weight minus birth weight. Wean End Daily Gain (called something different in methods) = Final weight minus weight at time of weaning.
These outcomes were statistically significant and suggest that PTCH1 can relate to weight gain, which is a significant indicator of influenza morbidity in the porcine population.
Discussion
The major findings of our present study demonstrate that the PTCH1 cell surface receptor is essential for infection by influenza virus and that targeted intervention against PTCH1, using either siRNA or antibodies, can decrease influenza infection. We also show that PTCH1 is broadly utilized by influenza during infection of cells from many different species, including humans, dogs and pigs. Finally, we translate the importance of these basic findings to a field study by demonstrating that PTCH1 relates to disease outcome during natural infection of porcine populations.
One novel aspect of our present study is the demonstration that PTCH1 regulates influenza infection of host cells. PTCH1 is the human homolog of the cellular receptor for sonic hedgehog, a secreted molecule that governs cellular organization, growth and survival during embryogenesis [11]. PTCH1 has also been implicated with roles as a tumor suppressor and is linked with various epithelial malignancies [10–12]. From a mechanistic standpoint, the basis of PTCH in influenza infection is unknown but the differential effect of antibody treatment at different MOIs may suggest a role for PTCH1 in relatively late stages of the viral life cycle. However, PTCH1 normally functions as a tumor suppressor, thus it will be important to consider the potential impact of targeted intervention against this molecule (e.g., using antibody therapies). Specifically, it may be necessary to limit PTCH1 targeting to acute indications. Such limitations may not preclude therapeutic applications since therapeutic intervention against influenza would generally be required for a limited time and this short duration of intervention might preclude undesired side effects.
Another important aspect of our present report is the utilization of RHGP to identify novel, host-directed targets for antiviral diseases. RHGP provides an opportunity to identify any target in a cell that regulates the phenotype under investigation, whether via overexpression or loss of expression [8, 13]. A central feature of RHGP is that one need not have prior knowledge of the target to determine its involvement in a disease process and our results further bolster this concept since PTCH1 had not been previously linked with influenza.
From the standpoint of disease management, there are interesting and important potential implications for our present findings. First, PTCH1 may provide an opportunity to intervene against infection by influenza using antibody or other approaches that target PTCH1 expression or function. The fact that comparable findings were obtained using multiple species (human, dog, pig) may indicate that PTCH1 represents an important core mechanism used for influenza infection. A central tenet of our host-directed approach to target infectious disease focuses on the idea that identification of fundamental pathways could provide a means to target multiple virus strains and future investigation should detail further the role of PTCH1 in different seasonal and pandemic forms of this viral pathogen [8]. A second opportunity arising from host-directed therapies is the concept that viruses may not have developed alternatives to important host mechanisms. If correct, then effective targeting of PTCH1 might provide an opportunity to overcome issues associated with the acquisition of drug resistance that have long plagued the treatment of influenza.
References
- 1.Thompson WW, Shay DK, Weintraub E, Brammer L, Cox N, Anderson LJ, Fukuda K. Mortality associated with influenza and respiratory syncytial virus in the United States. Jama-Journal of the American Medical Association. 2003;289:179–186. doi: 10.1001/jama.289.2.179. [DOI] [PubMed] [Google Scholar]
- 2.Belongia E, Kieke B, Coleman L, Donahue J, Irving S, Meece J, Vandermause M, Shay D, Gargiullo P, Balish A, Foust A, Guo L, Lindstrom S, Xu X, Klimov A, Bresee J, Cox N. Interim within-season estimate of the effectiveness of trivalent inactivated influenza vaccine - Marshfield, Wisconsin, 2007–08 influenza season (Reprinted from vol 57, pg 393–398, 2008) Jama-Journal of the American Medical Association. 2008;299:2381–2384. [Google Scholar]
- 3.Hayden F, Klimov A, Tashiro M, Hay A, Monto A, McKimm-Breschkin J, Macken C, Hampson A, Webster RG, Amyard M, Zambon M. Neuraminidase inhibitor susceptibility network position statement: antiviral resistance in influenza A/H5N1 viruses (vol 10, pg 873, 2005) Antiviral Therapy. 2006;11:130–130. [PubMed] [Google Scholar]
- 4.Luscher-Mattli M. Influenza chemotherapy: a review of the present state of art and of new drugs in development. Archives of Virology. 2000;145:2233–2248. doi: 10.1007/s007050070017. [DOI] [PubMed] [Google Scholar]
- 5.Fox JL. Antivirals become a broader enterprise. Nature Biotechnology. 2007;25:1395–1402. doi: 10.1038/nbt1207-1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tan SL, Ganji G, Paeper B, Proll S, Katze MG. Systems biology and the host response to viral infection. Nature Biotechnology. 2007;25:1383–1389. doi: 10.1038/nbt1207-1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lamb RA, Takeda M. Death by influenza virus protein. Nature Medicine. 2001;7:1286–1288. doi: 10.1038/nm1201-1286. [DOI] [PubMed] [Google Scholar]
- 8.Sui B, Bamba D, Weng K, Ung-Medoff H, Van Dyke J, Goldblatt M, Duan R, Kinch MS, Li W-L. The Use of Random Homozygous Gene Perturbation to Identify Novel Host-Oriented Targets for InfluenzaThe Use of Random Homozygous Gene Perturbation to Identify Novel Host-Oriented Targets for Influenza. Virology. 2009 doi: 10.1016/j.virol.2009.02.046. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ge Q, McManus MT, Nguyen T, Shen CH, Sharp PA, Eisen HN, Chen JZ. RNA interference of influenza virus production by directly targeting rnRNA for degradation and indirectly inhibiting all viral RNA transcription. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:2718–2723. doi: 10.1073/pnas.0437841100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Toftgard R. Hedgehog signalling in cancer. Cellular and Molecular Life Sciences. 2000;57:1720–1731. doi: 10.1007/PL00000654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lindstrom E, Shimokawa T, Toftgard R, Zaphiropoulos PG. PTCH mutations: Distribution and analyses. Human Mutation. 2006;27:215–219. doi: 10.1002/humu.20296. [DOI] [PubMed] [Google Scholar]
- 12.Katoh Y, Katoh M. Hedgehog signaling pathway and gastric cancer. Cancer Biology & Therapy. 2005;4:1050–1054. doi: 10.4161/cbt.4.10.2184. [DOI] [PubMed] [Google Scholar]
- 13.Reiske H, Sui B, Ung H, Donahue R, Li W-L, Goldblatt M, Li L, Kinch MS. Identification of Annexin A13 as a Regulator of Chemotherapy Resistance Using Random Homozygous Gene Perturbation. Analytical Quantitative Cytology Histology. 2009 In Press. [PubMed] [Google Scholar]