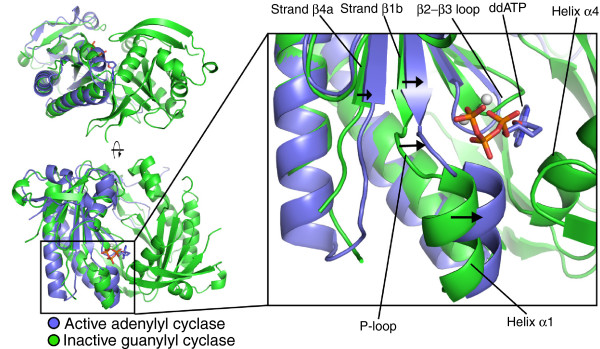
Figure 1.
The nucleotide cyclase wreath-like structure and active-site conformation. Comparison of the C. reinhardtii guanylyl cyclase catalytic core (green) with that of the C1:C2 domains of activated mammalian adenylyl cyclase (PDB ID: 1CJU; blue) bound to a nucleotide analog (shown as a stick model). The two active sites of the guanylyl cyclase are found in a cleft formed at the interface of two identical subunits (top left). The two subunits form a wreath-like structure, with active sites located at either end of the cleft (bottom left). The nucleotide analog marks the position of the single functional adenylyl cyclase active site. The guanylyl cyclase structure is presumed to be in an inactive conformation: in the active state of the nucleotide cyclases, it is expected that helix α1 (labeled in the right-hand panel) moves into the active site and toward helix α4 of the other subunit of the dimer. The arrows in the right-hand panel indicate the conformational changes required to generate the active configuration. From Winger et al. [3] Figure 4 (PDB 3ET6).