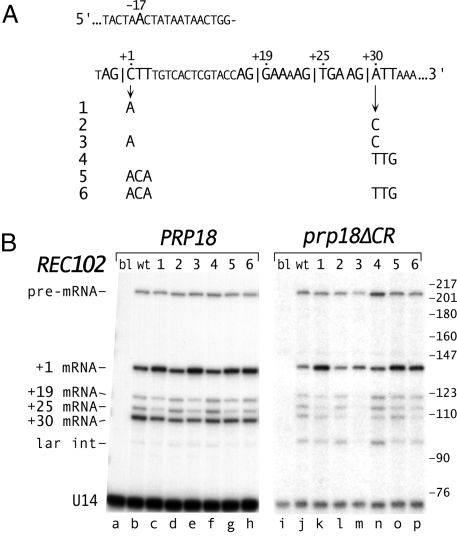
Fig. 5.
Effect of exon bases at the splice junctions on REC102 splice site choice. (A) Sequence of REC102 including its 3′ splice sites and branch site. Six mutations at the exon junctions are shown. (B) Primer extension analysis of splicing of REC102. The mutants of REC102 (blank, wt and 1–6) and the PRP18 ΔUPF1 (lanes a–h) and prp18ΔCR ΔUPF1 (lanes i–p) yeast are indicated at the top. The positions of the pre-mRNA, 4 mRNAs (numbers denote the first base of exon 2 in the spliced product), and lariat intermediate are indicated. Primer extension products were annealed to an oligo within exon1 and cut with HpaII before electorphoresis to facilitate separation (Fig. S5). To identify the primer extension products, RT-PCR products (using the original primer and a primer in exon 1) were purified from denaturing polyacrylamide gels and sequenced. The lariat intermediate was identified by treating the RNA from prp18ΔCR yeast with Dbr1 before primer extension (Fig. S5). The lengths of the primer extension products agree with their calculated values. B was assembled from two separate primer extension experiments. In direct comparisons, the amounts of spliced products in prp18ΔCR yeast were two- to fourfold less than in wild-type yeast.