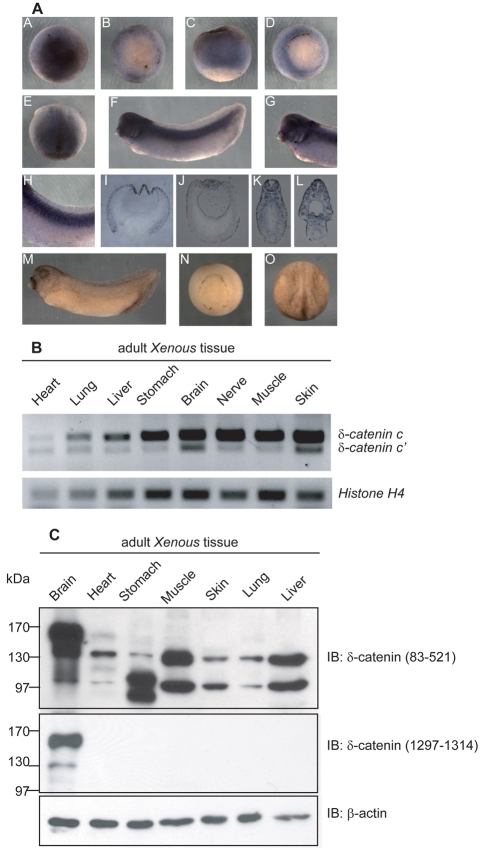
Fig. 3.
Spatial expression of Xenopus δ-catenin. (A) As viewed in animal versus vegetal regions, whole-mount in situ RNA hybridization detects δ-catenin mRNA signals in the ectoderm regions of blastula (subpanels A-C) and gastrula (subpanel D) embryos. At neurulation (subpanel E), the anterior and dorsal neural regions displayed the most apparent signals. Embryos at tadpole stages (subpanel F) showed a distinctive staining pattern in tissues of neural derivation such as brain, eye vesicle, ear vesicle, branchial arches (higher magnification in subpanel G) and spinal cord as well as somites (higher magnification in subpanel H). Subpanels I-L are cross-section views of paraffin-fixed embryos from corresponding stages. Sense probe hybridization was processed in parallel as negative controls (subpanels M-O). (B) RT-PCR analyses detect δ-catenin transcripts in all adult Xenopus tissues examined, with stronger expression in brain, nerve, muscle and skin. (C) Immunoblotting using an N-terminus-directed antibody detected three δ-catenin isoforms migrating at approximately 160, 130 and 100 kDa. The 130 and 100 kDa isoforms are ubiquitously present, whereas the 160 kDa appears to be brain specific. An antibody directed against the δ-catenin C-terminus reacts with the 160 kDa and 130 kDa isoforms in brain.