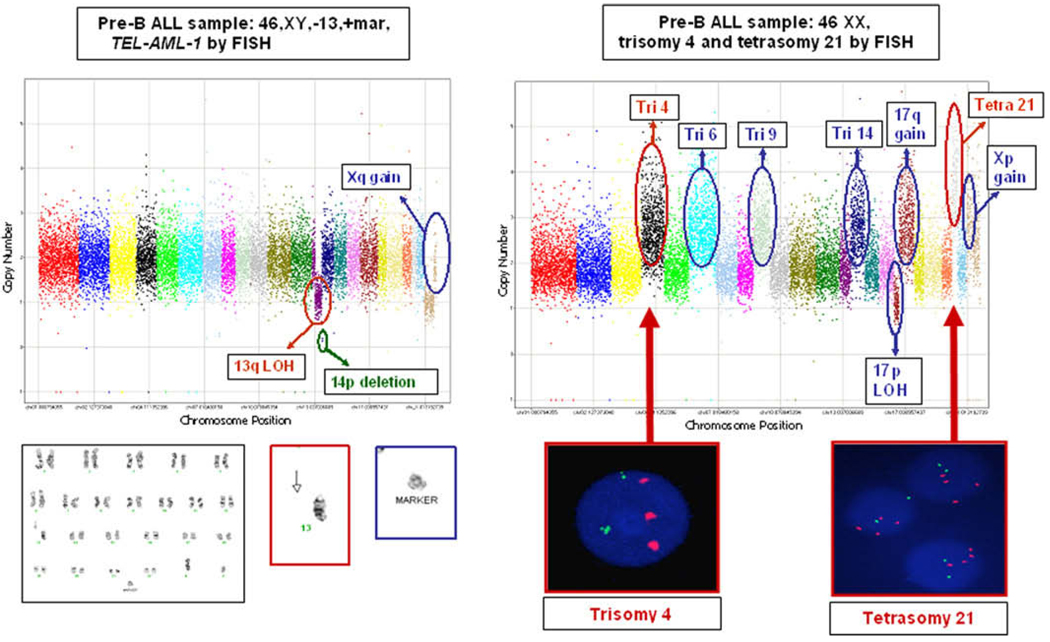
Fig. 1.
Molecular inversion probe analysis of pre-B ALL samples with customized 24 k cancer panel reveals additional allelic imbalance, compared with conventional karyotyping or fluorescence in situ hybridization (FISH) analysis. Chromosomes 1–22 and X are sequentially labeled in color, with copy number (linear scale) along the y-axis. (A) Karyotype 46,XY,−13, +mar; ETV6/RUNX1 fusion detected with TEL-AML1 FISH probe. The MIPs identified further allelic imbalance information (circled in green and blue) not described in the clinical karyotype. The chromosome 13 deletion (circled in red) was described as an entire deletion in the karyotype (red box, below graph), but was identified by MIPs as an isolated 13q deletion (13p remains intact). The unidentified marker (blue box, below graph) likely represents gain in Xq. (B) Karyotype 46,XX; trisomy 4 and tetrasomy 21 detected with FISH analysis. The MIPs identified further allelic imbalance information (circled in blue) not detected with karyotyping or FISH analysis and validate FISH findings (red boxes, below graph) of trisomy 4 and tetrasomy 21 (circled in red). Abbreviations: LOH, loss of heterozygosity; Tetra, tetrasomy; Tri, trisomy.