Figure 4.
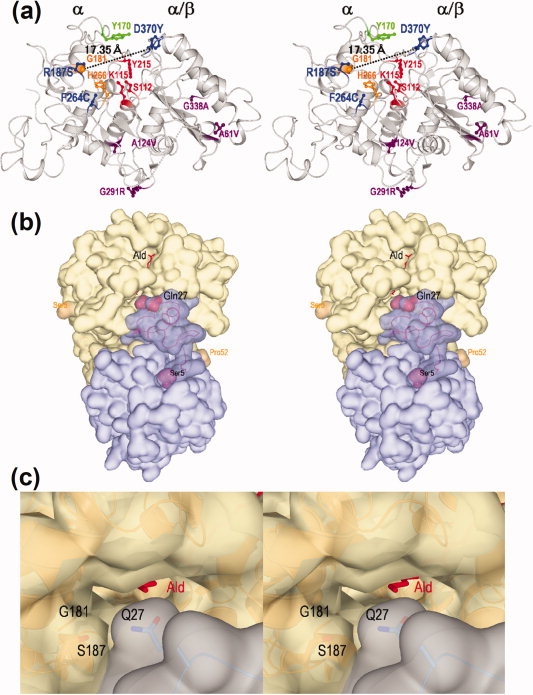
Stereo views of the three-dimensional structure of nylon oligomer hydrolase. (a) Overall structure of a monomer molecule of Hyb-S4M94 shown as a ribbon diagram. The left portion of the molecule is assigned as an α domain and the right portion as an α/β domain. The main chain folding is shown as a gray ribbon diagram, and the side-chains of the following amino acid residues are shown as a stick diagram. Proposed amino acid residues constituting the catalytic centers (S112/K115/Y215) are shown in red. Tyr170 is essential for Ald-hydrolytic activity is shown in green. Two alterations (G181D/H266N) increasing the Ald-hydrolytic activity in Hyb-24DN are shown in orange. Among the seven alterations differing between Hyb-24 and Hyb-S4M94, three (R187S/F264C/D370Y) were able to effectively increase the Ald-hydrolytic activity and are shown in blue, whereas the remaining four (A61V, A124V, G291R, and G338A) are shown in purple. (b) A homo-dimeric molecule of Hyb-S4M94 and interactions with substrate Ald. Surface structures of each monomeric unit are colored blue and gray, and substrate Ald is shown as a stick diagram colored red. Regions 1–4 and 53–57 were not traced because of poor electron density maps. The Ser5-Pro52 region located at the interface with another subunit is shown as semitransparent dark blue. Gln27, responsible for substrate binding, and Ser5/Pro52 are represented as a space-filing model. (c) Surface structure of the entrance of the catalytic cleft of the Hyb-S4M94-A112/Ald complex (closed form). The positions of Gln27, Gly181, and Ser187 are shown.
