Figure 5.
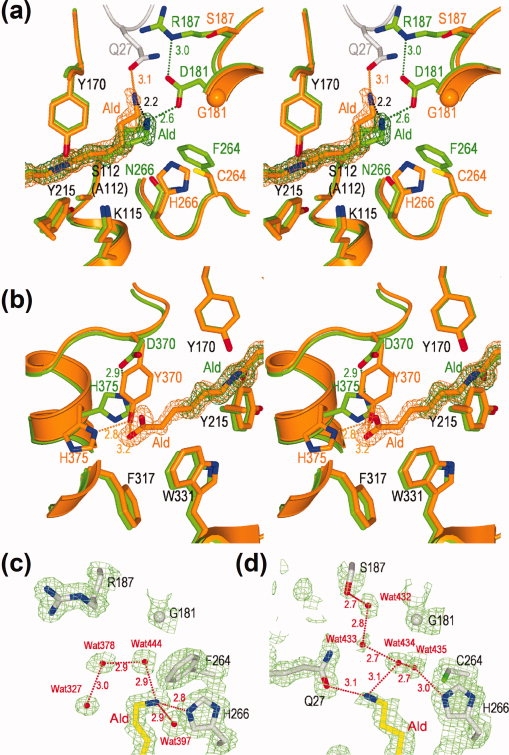
Interaction of Hyb-24 mutant enzymes with substrate Ald. (a,b) Stereo views of the superimposition of the Hyb-S4M94-A112/Ald complex (orange) with the Hyb-24DN-A112/Ald complex (green) at the amino-terminal portion (a) and carboxyl-terminal portion (b) of the substrate. The side-chains of catalytic/binding residues and substrate Ald with the 2Fo-Fc electron density maps contoured at 1.0σ are shown. (c,d) 2Fo−Fc electron density maps of the Hyb-24Y-A112/Ald complex (c) and the Hyb-S4M94-A112/Ald complex (d) contoured at 1.0σ. The side-chains of some residues [Gly181, Arg187 (Ser187), Phe264 (Cys264), His266, substrate Ald, and water molecules (Wat327, Wat378, Wat397, Wat432, Wat433, Wat434, Wat435, and Wat444) are shown. Hydrogen bonds between two atoms in the enzyme/substrate complex are indicated as dotted lines with the distance in angstroms.
