Figure 1.
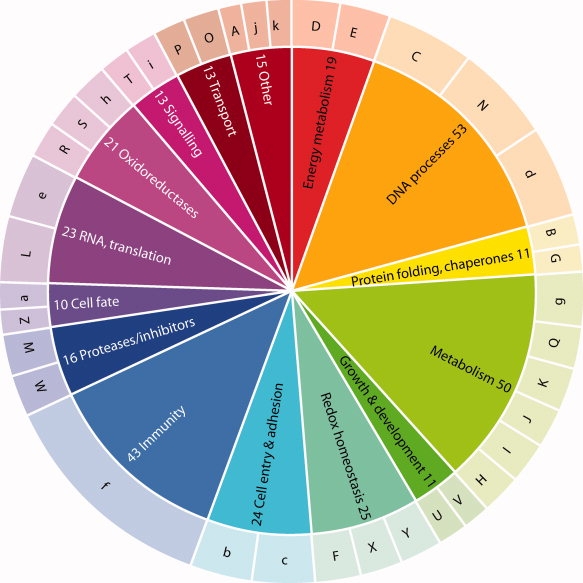
Functional supergroups in the Redox Pairs dataset (inner circle). The outer circle corresponds to groups in Supporting Information Table 1. Key to Groups: A, ion channel, ion pump modifier; B, protein folding, isomerization; C, cell cycle; D, Calvin cycle; E, photosynthesis, respiration, transport, maturation of proteins involved in these processes; F, destruction of oxygen radicals; G, redox metal homeostasis; H, amino acid metabolism; I, sulfur metabolism; J, nucleotide metabolism/homeostasis; K, isoprenoid biosynthesis; L, RNA-interacting; M, proteases; N, DNA repair; O, actin-related; P, carrier proteins; Q, other metabolism; R, NADP+-dependent oxidoreductase; S, other oxidoreductase; T, signaling; U, development; V, cell growth pathways; W, enzyme inhibitors; X, redox homeostasis; Y, ROS generating; Z, apoptosis; a, protein fate; b, cell entry; c, cell adhesion; d, DNA-interacting proteins; e, translation; f, immunity; g, carbohydrate metabolism; h, NAD+-dependent oxidoreductases; i, Ca handling; j, methyltransferase; k, other.
