Table 1. Data-collection and refinement statistics.
Values in parentheses are for the highest resolution bin.
| Data collection | |
| Resolution (Å) | 39.2–1.9 (2.0–1.9) |
| Completeness (%) | 99.6 (100) |
| Average redundancy | 4.9 (4.9) |
| I/σ(I) | 7.7 (1.9) |
| Rmerge† | 0.072 (0.386) |
| Space group | P21212 |
| Unit-cell parameters (Å, °) | a = 65.5, b = 85.4, c = 59.7, α = β = γ = 90.0 |
| Reflections | 26897 (3897) |
| Wilson B factor (Å2) | 22.9 |
| Refinement statistics | |
| Rwork | 0.184 (0.234) |
| Rfree | 0.199 (0.303) |
| Rwork reflections | 25546 |
| Rfree reflections | 1351 |
| No. of atoms | |
| Protein | 2567 [335 residues] |
| PLP | 16 |
| Solvent | 281 |
| Average B factors (Å2) | |
| Protein | 22.7 |
| PLP | 15.0 |
| Solvent | 31.1 |
| R.m.s.d. from ideal geometry‡ | |
| Bond lengths (Å) | 0.011 |
| Bond angles (°) | 1.4 |
| Ramachandran plot (%) | |
| Favored | 97.6 |
| Outliers | 0.0 |
| PDB code | 3ht5 |
R
merge = 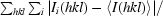
 .
.
Engh & Huber (1991 ▶).
