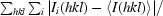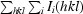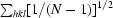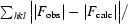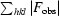Table 1. Data-collection and refinement statistics.
Values in parentheses are for the outermost shell of the resolution range.
| Data collection | |
| Space group | P212121 |
| Unit-cell parameters () | a = 78.3, b = 108.5, c = 148.3 |
| Resolution () | 63.411.45 (1.531.45) |
| R merge † | 0.077 (0.375) |
| R p.i.m. ‡ | 0.036 (0.172) |
| I/sdI | 11.6 (3.4) |
| Completeness (%) | 96.2 (89.5) |
| Redundancy | 5.1 (4.9) |
| Refinement | |
| No. of reflections | 1095072 (138933) |
| No. of unique reflections | 213226 (28642) |
| R work § | 0.167 (0.280) |
| R free | 0.189 (0.290) |
| No. of atoms | 10677 |
| Protein | 9336 |
| Ligands | 34 |
| Water | 1307 |
| Average B factors (2) | |
| Protein | 14.4 |
| Covalently bound pyruvate | 10.8 |
| Noncovalently bound pyruvate | 28.7 |
| PEG 400 | 50.0 |
| Waters | 16.5 |
| R.m.s. deviations¶ | |
| Bond lengths () | 0.012 |
| Bond angles () | 1.346 |
| Ramachandran statistics†† (%) | |
| Most favoured | 100 |
| Outliers | 0 |
| PDB code | 2wkj |