Table 2. Statistics for the quality of diffraction data sets for mutant p97.
All data sets were collected on SERCAT beamline ID22 at Argonne National Laboratory. Values in parentheses are for the last shell.
| Data set | R155H | R95G | R86A |
|---|---|---|---|
| Space group | R3† | P1 | P1 |
| Bound nucleotide | ATPγS | ATPγS | ATPγS |
| Unit-cell parameters (Å, °) | a = b = 134.2, c = 182.9† | a = 92.76, b = 103.3, c = 107.7, α = 97.7, β = 91.9, γ = 89.7 | a = 90.89, b = 102.6, c = 107.2, α = 97.5, β = 90.6, γ = 91.5 |
| Resolution (Å) | 50–2.1 (2.23–2.10) | 50–2.8 (2.90–2.80) | 50–2.85 (2.95–2.85) |
| Mosaicity (°) | 0.76 | 0.82 | 0.96 |
| No. of observations | 235711 | 151406 | 200205 |
| No. of unique reflections | 66557 | 87240 | 85163 |
| Completeness (%) | 99.5 (95.6) | 90.5 (71.5) | 91.8 (67.8) |
| Rmerge‡ | 0.057 (0.524) | 0.050 (0.493) | 0.049 (0.502) |
| I/σ(I) | 19.7 (1.7) | 12.3 (0.8) | 13.8 (1.02) |
| Phasing | |||
| Model used | PDB entry 1e32 | R155H N-D1 fragment | R155H N-D1 fragment |
| Log-likelihood gain | 446§ | 5792¶ | 6717¶ |
| R in initial rigid-body refinement | 0.43 | 0.38 | 0.37 |
| FOM | 0.36 | 0.59 | 0.62 |
R3 space group in hexagonal setting.
R
merge is defined as  −
− 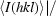
 , where I
i(hkl) is the intensity of the ith observation of a reflection with Miller indices hkl and 〈I(hkl)〉 is the mean intensity for all measured I(hkl) and Friedel pairs.
, where I
i(hkl) is the intensity of the ith observation of a reflection with Miller indices hkl and 〈I(hkl)〉 is the mean intensity for all measured I(hkl) and Friedel pairs.
MR was performed with Phaser using the automatic MR option. Two PDB files were provided: one for the D1-domain and another for the N-domain. We asked for two copies each of the D1-domain and N-domain. A single solution was obtained. A search with an intact N-D1 from PDB entry 1e32 produced a wrong solution with a log-likelihood gain of −42.
A monomeric N-D1 fragment from the R155H mutant structure was used as a phasing model, asking for six N-D1 subunits in a crystallographic asymmetric unit. A single solution was obtained.
