Figure 1.
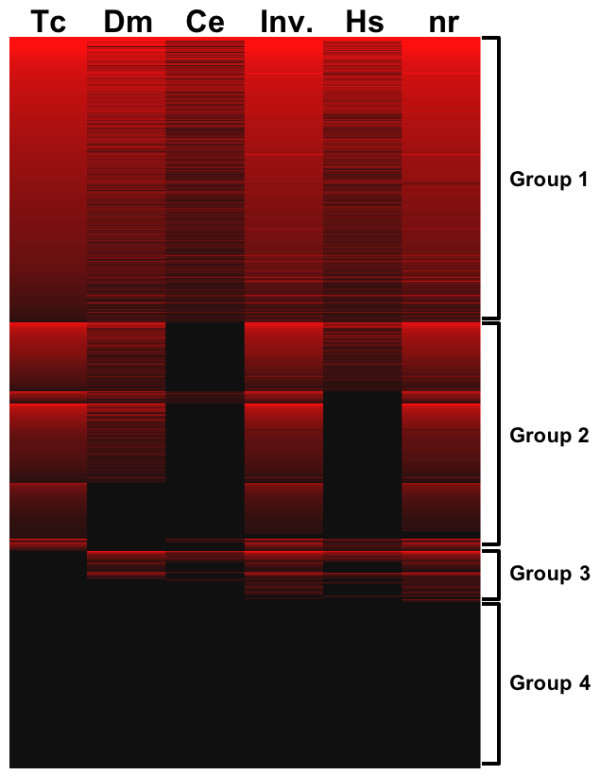
Overall comparison of O. taurus sequences with other protein datasets. Filtering and clustering analysis of assembled O. taurus ESTs based on BLASTx. Shown are bit scores against protein sequences from Tribolium castaneum (Tc, NCBI), Drosophila melanogaster (Dm, FlyBase), Caenorhabditis elegans (Ce, Sanger), invertebrate proteins (inv., NCBI), Homo sapiens (Hs, Ensembl), and non-redundant protein dataset (nr, NCBI). Each row represents a single Onthophagus sequence, and each column represents sequence matches to proteins from the indicated datasets, where the color intensity is proportional to the bit score (0 = black to 789 = brightest red). The Onthophagus sequences are grouped (Groups 1-4) according to the patterns of BLASTx sequence matches with proteins in the various datasets (E-value cut-off = 1 × 10-5), and clustered according to the bit scores. There are 1,086, 868, 194, and 633 sequences in groups 1-4, respectively. The complete dataset for this figure is available as Additional file 3.
