Figure 3.
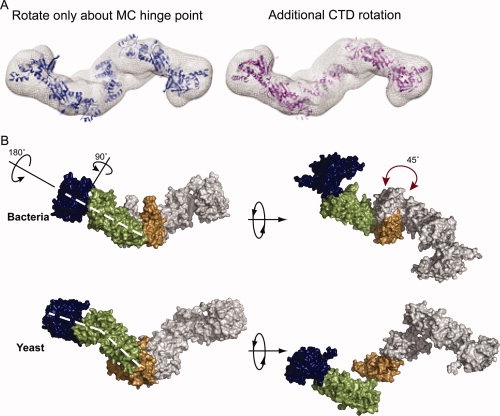
The apo yeast structure is distinct from the extended bacterial solution state. (A) When conformational space is searched only by rotations and translations about the hinge point, the resulting model loses the MC interace (blue). Rotation of the CTDs around the twofold axis by ∼45° (pink) restores the MD/CTD connection and better fits the Gasbor density (mesh). (B) In the yeast structure, the NTD is maximally extended by a ∼90° rotation away from the MD. The NM domains are also rotated away from one another by ∼180° as shown. These rotations cause a bend in the arms of the yeast protein not seen in HtpG (denoted by the dashed white lines). A rotation of the CTD (shown by the red arrow) causes a change in the point of contact between the MD and CTD leading to an angular chair-like structure.
