Figure 4.
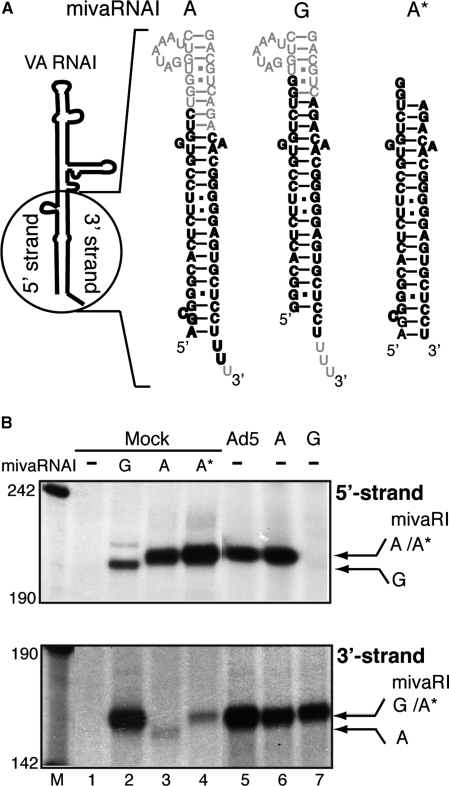
Mapping the Dicer processing site in VA RNAI(A) and VA RNAI(G). (A) Schematic drawing showing the difference in nucleotide sequence at the terminal stems of VA RNAI(A) and VA RNAI(G). The synthetic short double-stranded RNAs (shown in bold letters) were designed to mimic the predicted ‘A’ start and ‘G’ start mivaRNAI. Note that the structure of the extra base pairs at the beginning of the stem in the VA RNAI(A) structure were predicted. (B) S15 cytoplasmic extracts prepared from uninfected 293-Ago2 cells (Mock, lanes 1–4) or cells infected with wild-type Ad5 (lane 5) or cells transfected with plasmids pVA RNAI(A) (lane 6) or pVA RNAI(G) (lane 7) were assayed for RISC activity against the reverse VAI 5′ (upper panel) or reverse VAI 3′ (lower panel) target RNAs. The uninfected cytoplasmic extracts were mixed with water (lane 1), 2 pmol synthetic ‘G’ start mivaRNAI (lane 2), 2 pmol short version (lane 3) or the experimentally verified version of the ‘A’ start mivaRNAI (lane 4). The cleavage products from different mivaRNAIs are indicated by arrows.