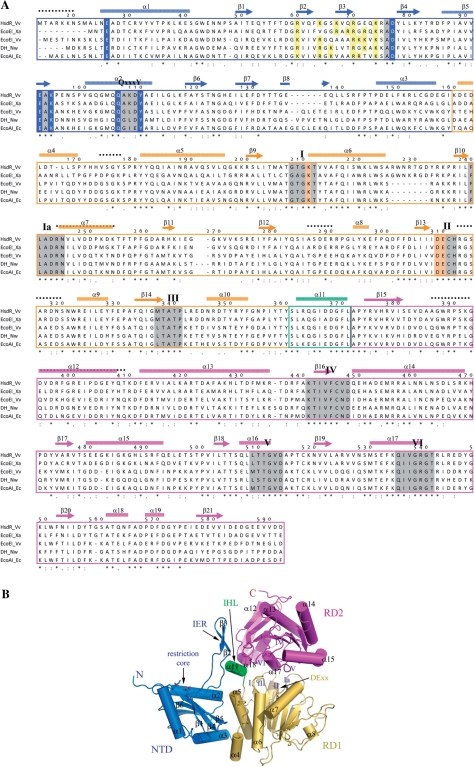
Figure 1.
Structure of HsdR_Vv. (A) Sequence comparison of HsdRs. Only the N-terminal fragment (Met14-Asp592) is displayed. The cylinders (α) and arrows (β) above the aligned sequences stand for the helix and strand, respectively. The numbering scheme follows the amino acid sequence of HsdR_Vv. Each domain for NTD, RD1 and RD2 is differentiated by colors. The catalytic residues at the NTD and the ATPase sites of RD1 are presented in blue and bright orange colored backgrounds, respectively. The conserved helicase-forming sequence motifs within the two RDs and the Q×××Y motif are presented in grey and green backgrounds, respectively. The regions of weak electron density in the present crystal structure are displayed by continuous black-dotted lines above the aligned sequences. Identical residues are marked by ‘asterisk’ and conserved residues by ‘:’ or ‘.’. Abbreviations used here: DH for DEAD-box containing helicase, Vv for V. vulnificus YJ016, Nw for Nitrobacter winogradskyi, Xa for Xanthobacter autotrophicus and Eco for E. coli. (B) Overall shape of the N-terminal fragment of HsdR_Vv structure. Each domain is displayed by ribbon diagrams and labeled using the same colors as in (A). The critical residues responsible for the restriction (NTD) and ATPase activities (RD1) are drawn by stick models. The spatial position of the conserved helicase-forming sequence motifs on both RDs are marked by Roman figures (I–VI). The figures, except for Figure 1A, were prepared by the PyMol Molecular graphics program of Delano Scientifics.