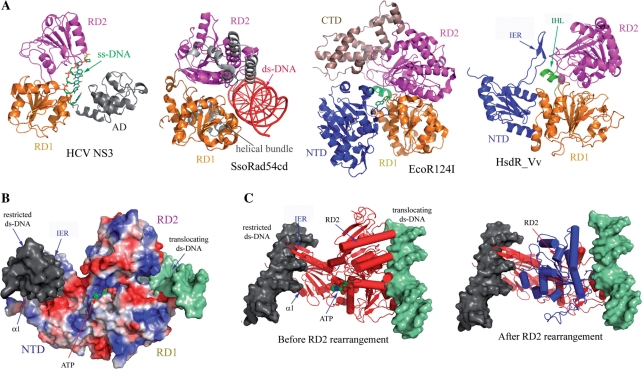
Figure 5.
Proposed translocation mechanism of HsdR_Vv. (A) HsdR_Vv is compared with ss-DNA translocator (HCV NS3), ds-DNA translocator (SsoRad54cd) and type I EcoR124I translocator (EcoR124I). The C-terminal helical bundle domain of EcoR124I was labeled by CTD. Note the remarkable structural difference in the relative spatial orientation of RD2 in HCV helicase and SsoRad54cd, which might imply the translocation mechanism of HsdR_Vv. (B) Modeling of ATP and two ds-DNAs at the NTD and at the surface along the two RDs. The N-terminal fragment structure of HsdR_Vv is represented by the surface potential map. The restricted (grey surface) and translocating (shallow green surface) ds-DNAs of HsdR_Vv were modeled by superposing the respective DNA complex structure of NgoMIV type II enzyme and SsoRad54cd ds-DNA translocator with the fragment structure of HsdR_Vv. The modeled ATP is displayed by the space filling model. In this model, the α1 helix of the NTD was straightened up to fit into the DNA groove as in the NgoMIV-DNA complex structure, while the IER (extended region) within the NTD was rotated towards the ds-DNA at the nucleolytic site, thereby locating the two stranded β-sheet into the DNA major groove. (C) Relative reorientation of RD2 in HsdR_Vv upon ATP and ds-DNA binding to RDs. The RD2 in the present structure is superposed well with the ss-DNA translocator HCV NS3 (before RD2 rearrangement), while RD2 is rearranged (blue in ‘After RD2 rearrangement’) when it is compared with the ds-translocator SsoRad54cd (After RD2 rearrangement).