Abstract
Restriction enzymes Ecl18kI, PspGI and EcoRII-C, specific for interrupted 5-bp target sequences, flip the central base pair of these sequences into their protein pockets to facilitate sequence recognition and adjust the DNA cleavage pattern. We have used time-resolved fluorescence spectroscopy of 2-aminopurine-labelled DNA in complex with each of these enzymes in solution to explore the nucleotide flipping mechanism and to obtain a detailed picture of the molecular environment of the extrahelical bases. We also report the first study of the 7-bp cutter, PfoI, whose recognition sequence (T/CCNGGA) overlaps with that of the Ecl18kI-type enzymes, and for which the crystal structure is unknown. The time-resolved fluorescence experiments reveal that PfoI also uses base flipping as part of its DNA recognition mechanism and that the extrahelical bases are captured by PfoI in binding pockets whose structures are quite different to those of the structurally characterized enzymes Ecl18kI, PspGI and EcoRII-C. The fluorescence decay parameters of all the enzyme-DNA complexes are interpreted to provide insight into the mechanisms used by these four restriction enzymes to flip and recognize bases and the relationship between nucleotide flipping and DNA cleavage.
INTRODUCTION
The complete rotation of a nucleotide from the DNA duplex, around the phosphate backbone, and into the catalytic pocket of an enzyme is commonly known as nucleotide flipping. It was first observed for the methyltransferase enzyme HhaI (1) and has since been observed in many other DNA methyltransferase systems (2–6) and several DNA-repair enzyme complexes (7,8). Recently, it was demonstrated that the Ecl18kI restriction enzyme uses nucleotide flipping as part of its DNA recognition mechanism (9). The co-crystal structure of Ecl18kI-bound DNA showed that the enzyme binds to the DNA and that both nucleotides that constitute the central base pair in its 5′-CCNGG-3′ (where N is any base) recognition sequence are flipped from the duplex, into the enzyme binding pockets. This report was the first example of an enzyme using nucleotide flipping in a situation where the flipped base is not the target of some chemical modification by the enzyme. This seemed a surprising and energetically extravagant method for recognizing a short-DNA sequence, but was soon found to apply to the evolutionarily related enzymes, PspGI (6,10–14) and EcoRII-C (11,15) the catalytic subunit of EcoRII (Table 1).
Table 1.
Restriction enzymes and their recognition sequences
| Enzyme | Recognition sequence |
|---|---|
| Ecl18kI | ↓CCNGG |
| EcoRII | ↓CCWGG |
| PspGI | ↓CCWGG |
| PfoI | T↓CCNGGA |
W denotes either A or T and downward arrows show the site targeted for phosphodiester cleavage. The common CCGG interrupted tetranucleotide is indicated in boldface.
Recently, using the fluorescent analogue of adenine, 2-aminopurine (2AP), we demonstrated that Ecl18kI, EcoRII-C and PspGI unstack bases at the centre of their recognition sequences in solution (11). The increase in 2AP fluorescence intensity upon complex formation varied significantly in the Ecl18kI, PspGI and EcoRII-C complexes. The 2AP fluorescence increase was largest for PspGI (64-fold), intermediate for EcoRII-C (∼12-fold) and smallest for Ecl18kI (6.5-fold).
Although an increase in steady-state intensity is useful as an indicator of destacking of 2AP from the duplex, the magnitude of the intensity change is difficult to interpret, since different distributions of the 2AP population amongst the various conformational states of the duplex-enzyme complex may give similar steady-state fluorescence intensities. Time-resolved fluorescence spectroscopy gives a more detailed picture of the environment around a fluorophore and the heterogeneity of the environment. A fluorophore in a homogeneous environment exhibits one lifetime; its fluorescence decay is monoexponential. The decay is multiexponential if the fluorophore is partitioned between several environments or conformations that provide distinctly different quenching efficiencies. Thus, 2AP-containing DNA duplexes typically show fluorescence decays described by four lifetime components, reflecting the existence of the duplex in a variety of conformational states. Changes in the 2AP fluorescence decay on enzyme binding can be interpreted to give a detailed picture of the conformational distortion that is induced, including an assessment of the extent of base flipping (16–19).
Here, we have used fluorescence spectroscopy to investigate the nucleotide flipping mechanism of the structurally characterized 5-bp cutters, Ecl18kI, EcoRII-C and PspGI, and the 7-bp cutter, PfoI, for which the crystal structure is unknown. Steady-state fluorescence experiments indicate that the central base pair is unstacked in the PfoI-DNA complex, resulting in ∼1000-fold increase of the 2AP fluorescence intensity, compared with the free DNA duplex. Time-resolved fluorescence experiments give new insight into the conformational dynamics of base flipping and the nature of the interactions between the extrahelical 2AP bases and the enzymes. We seek to interpret this insight to address pertinent mechanistic questions regarding these novel base flipping systems, such as is there cooperativity between the enzyme monomers to produce stable base flips; what is the orientation (extent of rotation) of the flipped base relative to the DNA duplex in the solution phase and what is the relationship between base orientation and catalysis?
MATERIALS AND METHODS
Oligonucleotides
2AP-containing oligodeoxynucleotides were obtained from Integrated DNA Technologies (HPLC grade, Coralville, USA) and non-modified oligodeoxynucleotides were purchased from Metabion (HPLC grade, Martinsried, Germany). In order to assemble duplexes, appropriate oligodeoxynucleotides (Table 2) containing 2AP or non-fluorescent control strands were mixed with a 1.05-fold molecular excess of complementary strands in the reaction buffer A [33 mM Tris-acetate (pH 7.9 at 25°C), 66 mM potassium acetate], heated to 85°C and allowed to cool slowly over several hours at room temperature. For the DNA binding and cleavage studies, both strands of the 25-bp duplexes were 5′-end labelled with [γ-33P]ATP (Hartmann Analytic, Braunschweig, Germany) using a DNA labelling kit (Fermentas, Vilnius, Lithuania).
Table 2.
Oligoduplexes used in this studya
| Oligoduplex | Sequence |
|---|---|
| T/2a | 5′ CGCACGCCTTCCTGGAAGCACACTA 3′ |
| 3′ GCGTGCGGAAGG2CCTTCGTGTGAT 5′ | |
| 2/2a | 5′ CGCACGCCTTCC2GGAAGCACACTA 3′ |
| 3′ GCGTGCGGAAGG2CCTTCGTGTGAT 5′ | |
| 2NSb | 5′ CGCACGCCTTCCTGGAAGCACACTA 3′ |
| 3′ GCGTGCGGA2GGACCTTCGTGTGAT 5′ | |
| 2NS′b | 5′ CGCACGCCTTCCTGGAAGCACACTA 3′ |
| 3′ GCGTGCGG2AGGACCTTCGTGTGAT 5′ | |
| T/A | 5′ CGCACGCCTTCCTGGAAGCACACTA 3′ |
| 3′ GCGTGCGGAAGGACCTTCGTGTGAT 5′ |
aOverlapping Ecl18kI/EcoRII-C/PspGI/PfoI recognition site is in boldface; 2 = 2AP, is indicated in boldface and underlined.
bDuplexes 2NS and 2NS′ contain 2AP introduced immediately adjacent to the target site. The 2NS duplex was used in the Ecl18kI/EcoRII-C/PspGI experiment and the 2NS′ duplex was used in the PfoI experiment.
Strains, plasmids and proteins
Cloning of the PfoI restriction-modification system from Pseudomonas fluorescens biovar 126 will be described elsewhere. Recombinant wild-type (wt) pfoIR gene was cloned in the pBAD24 [ApR] vector. Strain DH10B (ara−) (F− mcrA Δ(mrr-hsdRMS-mcrBC) φ80lacZΔM15 ΔlacX74 recA1 endA1 deoR araD139 Δ(ara, leu)7697 galU galK16 galE15 l− rpsL nupG) containing plasmid pHSG415ts_M.Ecl18kI [CmR] (20) bearing the ecl18kIM gene was used as a host for transformation of [pBAD24_R.PfoI] [ApR] containing the pfoIR gene. To express the PfoI protein cells were grown in LB medium with appropriate antibiotics at 30°C to an OD600 of 0.7 and PfoI expression was induced by addition of arabinose to the final 0.2% concentration. The PfoI protein was purified as described previously (21). Fractions containing PfoI enzyme were pooled and dialysed against the storage buffer [10 mM Tris-HCl (pH 8.0 at 25°C), 300 mM KCl, 1 mM DTT, 1 mM EDTA (ethylenediaminetetraacetic acid), 50% glycerol] and stored at −20°C. The protein preparation was >90% pure according to a Coomassie-blue-stained sodium dodecyl sulphate (SDS)-gel.
The preparations of wt Ecl18kI, the W61Y and W61A variant of Ecl18kI, PspGI and EcoRII-C were carried out as described previously (13). All protein concentrations were determined from the absorption at 280 nm and refer to the dimers. An extinction coefficient 56 300 M−1 cm−1 was calculated for the PfoI dimer using the Vector NTITM software.
Gel mobility shift assay
33P-labelled DNA duplexes (Table 2) at 0.2 nM concentration were mixed with increasing amounts of protein in the presence of 5 mM Ca2+. Apparent Kd values were determined as described previously (22).
DNA cleavage activity
The DNA cleavage activities of PfoI were monitored using a 25-bp DNA duplex containing a 33P-label in both DNA strands (Table 2). Cleavage reactions were conducted at 15°C in the reaction buffer A containing 10 mM magnesium acetate and 0.1 mg ml−1 BSA (bovine serum albumin) using 100 nM of duplex and 1000 nM of protein. Aliquots were removed at timed intervals and quenched by mixing with loading dye [95% (v/v) formamide, 0.01% (w/v) bromophenol blue] before denaturing gel electrophoresis. The samples were analysed and quantified as described previously (23).
Steady-state fluorescence spectroscopy
PfoI steady-state fluorescence measurements were acquired in photon counting mode on a Fluoromax–3 (Jobin Yvon, Stanmore, UK) spectrofluorometer. Sample temperatures were maintained at 25°C by a circulating water bath. Emission spectra (340–420 nm) were recorded at an excitation wavelength λex = 320 nm with excitation and emission bandwidths of 2 and 8 nm, respectively. At least three scans were averaged for each spectrum. Emission spectra were collected in reaction buffer A in the presence of 10 mM calcium acetate using 250 nM DNA alone or 250 nM DNA mixed with a 5-fold excess of the protein to ensure complete binding of the DNA. Control spectra used for the background subtraction corrections were collected under identical conditions except that duplex T/A was used instead of the fluorescent DNA. For the DNA duplex titration experiment, emission spectra of the 250 nM DNA with protein in the range of 0–2000 nM were collected.
Enzymatic digestion of the DNA duplexes was done by addition of P1 nuclease (0.5 U, Sigma Aldrich, Taufkirchen) to a solution (200 μl) containing thermically denaturated 250 nM duplex T/2 or 2/2 in 10 mM Bis-Tris-HCl (pH 5.8 at 25°C), 100 mM NaCl buffer supplemented with 5 mM zinc acetate and incubating for 1–16 h at 37°C or 56°C.
Time-resolved fluorescence spectroscopy
In order to ensure complete binding of the DNA by restriction enzymes, samples were prepared containing 1 μM DNA duplex, 5 μM enzyme and 10 mM calcium acetate in reaction buffer A. For the time-resolved measurements of the DNA duplexes alone, 5 μM of DNA duplex in the same buffer was used.
Time-resolved fluorescence spectroscopy was performed using the technique of time-correlated single photon counting. The samples were measured in an Edinburgh Instruments spectrometer equipped with TCC900 photon counting electronics. The excitation source was a Ti-Sapphire femtosecond laser system (Coherent, 10 W Verdi and Mira Ti-Sapphire) producing pulses of ∼200 fs at 76 MHz repetition rate. The output of the Ti-Sapphire laser was passed through a pulse picker to reduce the repetition rate to 4.75 MHz and then frequency tripled to give an output at 316 nm. The emission from the sample was collected orthogonally to the excitation beam through a polarizer set at the magic angle (54.7°) with respect to the vertically polarized excitation. The fluorescence was passed through a monochromator (bandpass 10 nm) and detected by a Hamamatsu microchannel plate photomultiplier (R3809U-50). The instrument response of the system, measured using a dilute aqueous solution of Ludox (Sigma-Aldrich) was 70 ps full-width at half maximum.
Fluorescence decay curves were recorded on a 50 ns timescale, resolved into 4096 channels, to a total of 10 000 counts in the peak channel. Decay curves were analysed using a standard iterative reconvolution method, assuming a multiexponential decay function,
| 1 |
where Ai is the fractional amplitude and τi is the fluorescence lifetime of the i-th decay component.
Fluorescence was excited at 316 nm and decay curves were recorded at three emission wavelengths, 370, 380 and 390 nm. The three decays were analysed globally using Edinburgh Instruments ‘FAST’ software, i.e. they were fitted simultaneously, with lifetimes, τi, as common parameters. The quality of fit was judged on the basis of the reduced chi-square statistic, χ2, and the randomness of residuals.
RESULTS
Nucleotide flipping by PfoI
Restriction endonuclease PfoI from P. fluorescens biovar 126 recognizes the interrupted hexanucleotide palindromic sequence 5′-TCCNGGA-3′ and cuts phosphodiester bonds on the 5′-sides of the outer cytosines (21). DNA cleavage by PfoI produces the same protruding pentanucleotide 5′-ends as Ecl18kI and the EcoRII-C/PspGI enzymes (Table 1). The functional similarity of PfoI to the base flipping enzymes Ecl18kI, PspGI and EcoRII-C, prompted us to investigate whether PfoI flips central nucleotides from its recognition sequences, using the fluorescent adenine analogue, 2AP. Gel shift experiments revealed that the binding affinity of PfoI with duplex DNA is independent of the 2AP substitution at the site for possible base flipping (data not shown). In the buffer supplemented with Mg2+ ions, PfoI cleaved duplexes containing or lacking 2AP at identical rates (data not shown). All 2AP fluorescence experiments were performed in the presence of Ca2+ ions, which do not support catalysis but are required for stable, specific DNA binding for restriction enzymes (20,22,24).
The steady-state fluorescence intensity of the T/2 and 2/2 DNA duplexes increased dramatically when bound by PfoI, as shown in Figure 1A. The relative increase in intensity was much larger than for other base flipping restriction endonucleases and other flipping enzymes (25–37) and indicates that the 2AP is flipped from the duplex in the PfoI-DNA complex. When duplex 2/2 was bound by PfoI, the increase in fluorescence intensity was approximately double than that observed for T/2, implying that both target bases are flipped by PfoI, as seen previously for Ecl18kI, EcoRII-C and PspGI (11). Digestion of the T/2 (or 2/2) duplex with nuclease P1 to give the free 2AP deoxyribonucleotide, revealed that the intensity of the flipped 2AP in the PfoI complex was ∼80% of that of the free deoxyribonucleotide, as shown in Figure 1B.
Figure 1.
2AP fluorescence intensity in the ternary complexes with PfoI. (A) Bar chart comparing fluorescence intensity values of the corrected fluorescence emission spectra at fluorescence maximum (360 nm for EcoRII-C/PspGI/PfoI, 367 nm for wt Ecl18kI, 365 nm for Ecl18kI W61Y and W61A and 370 nm for T/2 duplex alone) in the restriction enzyme-T/2-Ca2+ ternary complexes. Reactions contained 1250 nM enzyme, 250 nM DNA duplex T/2 and 10 mM Ca2+ ions. The fluorescence intensity data for Ecl18kI, EcoRII-C and PspGI have been published previously (11,13). (B) Fluorescence emission spectra of free 250 nM duplex T/2, 250 nM T/2 duplex in the presence of saturating concentration of PfoI (1250 nM) and of 250 nM T/2 duplex after digestion with nuclease P1.
Time-resolved fluorescence of restriction enzyme-DNA complexes
To obtain a more detailed picture of the environment of the extrahelical bases, we carried out time-resolved fluorescence measurements on 2AP-labelled DNA in enzyme-DNA–Ca2+ ternary complexes. Recorded decays were modelled by the minimum number of lifetime components required to give an acceptable fit. In a simple interpretation, each lifetime (τi) can be considered to represent a distinct conformational state and its fractional amplitude (Ai) indicates the fraction of the population occupying this state. This simple model should not be regarded as an exact physical description of the ensemble of duplex conformations, since each lifetime is likely to represent a distribution of conformations in which 2AP experiences similar quenching rates. Nevertheless, it provides valuable insight into the conformational properties of the duplex and, more significantly in the present context, the changes in these properties induced by enzyme binding.
DNA duplex 2NS
Enzymatic binding to the 2NS duplex caused minimal perturbation to the fluorescence decay of the 2AP, as shown in Table 3. The small changes in decay parameters indicate a slight perturbation of base stacking in the duplexes on enzyme binding. As will be shown below, much greater effects are seen on enzyme binding to the duplexes with 2AP in the recognition sequence.
Table 3.
Fluorescence lifetimes, τi and corresponding fractional amplitudes, Ai, for the DNA duplex 2NS, where the 2AP lies outside the enzymatic recognition sequence, and its complexes with PfoI, PspGI, Ecl18kI and EcoRII-C
| Solution | τ1/ns | τ2/ns | τ3/ns | τ4/ns | A1 | A2 | A3 | A4 |
|---|---|---|---|---|---|---|---|---|
| 2NS | 0.02 | 0.30 | 2.1 | – | 0.98 | 0.01 | 0.01 | – |
| PfoI+2NS | 0.05 | 0.91 | 3.4 | 8.6 | 0.87 | 0.05 | 0.04 | 0.04 |
| PspGI+2NS | 0.05 | 0.49 | 2.6 | 9.4 | 0.92 | 0.05 | 0.02 | 0.01 |
| Ecl18kI+2NS | 0.04 | 0.44 | 2.3 | 7.9 | 0.94 | 0.02 | 0.03 | 0.02 |
| EcoRII-C+2NS | 0.05 | 0.50 | 2.8 | 8.5 | 0.88 | 0.06 | 0.04 | 0.02 |
DNA duplexes T2 and 2/2
The fluorescence decay parameters for the 2AP-labelled duplexes, T2 and 2/2 are shown in Table 4.
Table 4.
Fluorescence lifetimes, τi and corresponding fractional amplitudes, Ai, for duplexes T/2 and 2/2
| Solution | τ1/ns | τ2/ns | τ3/ns | τ4/ns | A1 | A2 | A3 | A4 |
|---|---|---|---|---|---|---|---|---|
| T/2 | 0.03 | 0.41 | 2.4 | 9.7 | 0.94 | 0.03 | 0.02 | 0.01 |
| 2/2 | 0.07 | 0.46 | 2.0 | 7.8 | 0.89 | 0.05 | 0.05 | 0.01 |
The fluorescence decays are described by four exponential components, as seen typically for 2AP-labelled DNA (16–18,38). The shortest lifetime (τ1 < 100 ps) is attributed to 2AP that is intrahelical and whose excited state is rapidly deactivated by electron transfer from guanine bases in close proximity (39,40). Approximately 90% of the free duplexes, both T/2 and 2/2, exist in this conformation. The longest decay component (τ4 > 7 ns) is attributed to 2AP that is extrahelical and experiencing a solvated environment where quenching of the fluorescence by electron transfer from guanine is not favourable. The value of the latter decay time is comparable with that found for free 2AP-riboside in solution, 10.6 ns (41,42). This component accounts for a tiny fraction (∼1%) of the population. The intermediate lifetime species represent imperfectly stacked duplex conformations in which 2AP is intrahelical, but cannot be readily quenched by electron transfer from guanine. The near absence of extrahelical conformations and the overwhelming predominance of highly quenched conformations indicate the 2AP base to be firmly stacked within the duplex, in both the cases. There are some differences in decay parameters between T/2 and 2/2 duplexes, although the intrastrand sequence context of 2AP is identical in both. Thus, the conformational properties are influenced by the different base pair strengths. Notably, τ1 is significantly shorter in T/2, indicating enhanced stacking of the 2AP with guanine neighbours.
PfoI
The fluorescence decay parameters of the ternary complexes of the duplexes, T/2 and 2/2, with PfoI and calcium differ dramatically from those of the free duplexes, as shown in Table 5.
Table 5.
Fluorescence lifetimes, τi and the corresponding fractional amplitudes, Ai, for the PfoI-DNA-Ca2+ ternary complexes
| Solution | τ1/ns | τ2/ns | τ3/ns | τ4/ns | A1 | A2 | A3 | A4 |
|---|---|---|---|---|---|---|---|---|
| PfoI+T/2 | – | – | 3.7 | 8.5 | – | – | 0.14 | 0.86 |
| PfoI+2/2 | – | – | 3.4 | 8.3 | – | – | 0.09 | 0.91 |
The fluorescence decays of the PfoI-T/2-Ca2+ and PfoI-2/2-Ca2+ complexes are essentially identical. Each can be well described by only two components with lifetimes of 3.7 (3.4) ns and 8.5 (8.3) ns. Hence, the 2AP is found in only two distinct environments in these complexes and >80% of the population is free from quenching interactions. This is consistent with the large increase in fluorescence intensity observed on enzyme binding. The complete absence of sub-nanosecond decay components indicate that all of the target bases are ejected from the duplex. The essentially identical decay parameters for both duplexes demonstrate that both target bases in duplex 2/2 are flipped and experience the same extrahelical environment. This is consistent with the observation that the fluorescence intensity for PfoI bound to 2/2 is double that observed when bound to T/2.
PspGI
The fluorescence decays of the PspGI-DNA-Ca2+ ternary complexes resemble those of the free duplexes, in being described by four exponential components. However, the values of the decay parameters are substantially different from those of the free duplexes, as shown in Table 6 and illustrated graphically in Figure 2.
Table 6.
Fluorescence lifetimes, τi and the corresponding fractional amplitudes, Ai, for the PspGI-DNA-Ca2+ ternary complexes
| Solution | τ1/ns | τ2/ns | τ3/ns | τ4/ns | A1 | A2 | A3 | A4 |
|---|---|---|---|---|---|---|---|---|
| PspGI+T/2 | 0.20 | 0.74 | 2.6 | 8.8 | 0.55 | 0.34 | 0.07 | 0.04 |
| PspGI+2/2 | 0.16 | 0.63 | 2.7 | 8.7 | 0.64 | 0.26 | 0.05 | 0.05 |
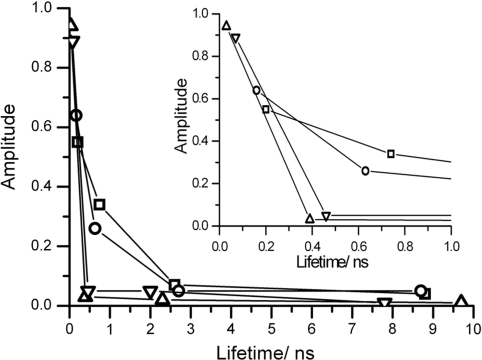
Figure 2.
The decay components of the free duplexes, free T/2 (triangles) and 2/2 (inverted triangles) and the ternary complexes, PspGI-T/2-Ca2+ (squares) and PspGI-2/2-Ca2+ (circles), plotted as fractional amplitude versus fluorescence lifetime. Inset shows the same data with an expanded x-axis to highlight changes in the two subnanosecond decay components on enzyme binding.
Binding of each duplex by PspGI results in a significant increase in the shortest lifetime, τ1, from 30 to 200 ps for the T/2 duplex, and from 70 to 160 ps for the 2/2 duplex. The amplitude of this shortest component also decreases significantly in both the cases. These changes are indicative of destacking of 2AP from the duplex. However, the molecular environment seen by 2AP in these complexes differs markedly from that in the PfoI complexes, with 90% of the population in highly quenched conformations, described by the two subnanosecond decay times, τ1 and τ2.
Ecl18kI
The decays of the duplexes complexed with wt Ecl18kI are also described by four exponential components, and are rather similar to those of the free duplexes. Nevertheless, as shown in Table 7 and illustrated graphically in Figure 3, there are significant differences between the decay parameters of the bound and free duplexes.
Table 7.
Fluorescence lifetimes, τi and the corresponding fractional amplitudes, Ai, for the ternary complexes of T/2 and 2/2 with wt Ecl18kI and its mutants W61Y and W61A
| Solution | τ1/ns | τ2/ns | τ3/ns | τ4/ns | A1 | A2 | A3 | A4 |
|---|---|---|---|---|---|---|---|---|
| Ecl18kI+T/2 | 0.07 | 0.77 | 3.1 | 9.0 | 0.74 | 0.09 | 0.11 | 0.06 |
| Ecl18kI+2/2 | 0.07 | 0.70 | 3.1 | 8.9 | 0.80 | 0.07 | 0.08 | 0.05 |
| W61Y+T/2 | 0.18 | 0.62 | 1.7 | 7.0 | 0.59 | 0.29 | 0.11 | 0.01 |
| W61Y+2/2 | 0.17 | 0.53 | 1.5 | 6.1 | 0.55 | 0.29 | 0.15 | 0.01 |
| W61A+T/2 | 0.14 | 0.91 | 3.8 | 9.2 | 0.61 | 0.12 | 0.08 | 0.19 |
| W61A+2/2 | 0.16 | 1.3 | 4.6 | 9.0 | 0.38 | 0.12 | 0.24 | 0.26 |
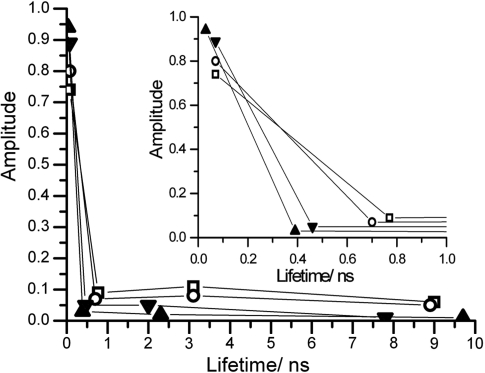
Figure 3.
Plot of fractional amplitude versus fluorescence lifetime for free T/2 (filled triangles) and 2/2 (filled inverted triangles) and the ternary complexes, Ecl18kI-T/2-Ca2+ (squares) and Ecl18kI-2/2-Ca2+ (circles). Inset shows magnified view of the subnanosecond lifetime range.
The decay parameters of the two ternary complexes are virtually identical. Around 75% of the ternary complex population shows a very short, 70 ps, lifetime characteristic of highly quenched 2AP. In comparison, in the free duplexes, a significantly greater fraction, 90%, of the 2AP population is highly quenched. The intermediate lifetimes (τ2 and τ3) increase significantly in the ternary complexes, compared with the free duplexes, as do their amplitudes. These changes in the decay parameters are clear indicators that the DNA duplex structure is perturbed by the binding of Ecl18kI, but are not typical of base flipping. To further investigate the possibility of base flipping, the tryptophan residue (W61) within the Ecl18kI flipping pocket was mutated to W61Y and W61A variants.
As shown in Table 7, both the amplitudes and lifetimes of the 2AP decays change when the W61 residue is mutated. The W61Y mutation leads to a significant increase in the shortest lifetime, τ1, and a decrease in its amplitude. There is a corresponding increase in the amplitude of component 2. The W61A mutation results in a general increase in lifetimes (except for the longest component). As for W61Y, the amplitude of the shortest lifetime component decreases appreciably with this mutation. For this mutant alone, the decay parameters of the ternary complexes containing the T/2 and 2/2 duplexes are quite different, particularly the amplitudes of components 1 and 3. This signifies a transfer of population from highly quenched to relatively unquenched conformations in the W61A-2/2-Ca2+ complex, compared with W61A-T/2-0Ca2+.
EcoRII-C
As shown in Table 8, the decay parameters of the EcoRII-C complexes are very similar to those of the wt Ecl18kI complexes (Table 7). The decays of the bound duplexes are virtually identical and 80% of the 2AP population has a very short, 80 ps, lifetime. As for Ecl18kI, there is clear evidence that duplex conformation in the vicinity of the target base is affected by binding of EcoRII-C, but the decay parameters do not conform to our expectations for base flipping.
Table 8.
Fluorescence lifetimes, τi and the corresponding fractional amplitudes, Ai, for the ternary complexes of T/2 and 2/2 with EcoRII-C
| Solution | τ1/ns | τ2/ns | τ3/ns | τ4/ns | A1 | A2 | A3 | A4 |
|---|---|---|---|---|---|---|---|---|
| EcoRII-C+T/2 | 0.08 | 0.53 | 2.5 | 8.5 | 0.81 | 0.12 | 0.04 | 0.03 |
| EcoRII-C+2/2 | 0.08 | 0.56 | 2.7 | 8.4 | 0.82 | 0.10 | 0.05 | 0.03 |
DISCUSSION
Comparison of steady-state and time-resolved measurements
We note here a significant discrepancy between the fluorescence enhancement seen in the steady-state data and that which is readily calculated from the time-resolved fluorescence measurements (Supplementary Data). Figure 1 shows a fluorescence enhancement of the order of 1000-fold for base flipping (on the T/2 duplex) by PfoI. However, from the time-resolved data, we would predict an increase in the steady-state intensity of only ∼50-fold. The discrepancy is not so large for the 2/2 duplex, which shows a steady-state intensity increase of ∼130-fold and a predicted increase of ∼30-fold from the time-resolved results. The disagreement between the two measurements can be explained by the occurrence of extremely rapid quenching of some fraction of the excited state 2AP that is stacked within the free DNA duplex, such that it decays on a timescale that is faster than the time resolution of our experiments and has a vanishingly small quantum yield. Hence, there is a population of non-fluorescent 2AP in the free duplexes whose quantum yields increase dramatically upon base flipping. Effectively, the concentration of ‘fluorescent’ 2AP is increased by base flipping. This has a dramatic effect on the steady-state intensity, but is not apparent in the time-resolved measurements, since the decay parameters are independent of the concentration of emitting molecules. Previous ultrafast spectroscopic measurements of 2AP in DNA demonstrate the existence of quenching on the picoseconds timescale (43,44). The better agreement between steady-state and time-resolved measurements on the 2/2 duplex implies a smaller population of ultra-quenched 2AP which is consistent with the mis-pairing of 2AP in this duplex.
Nucleotide flipping by PfoI
Previous time-resolved fluorescence studies of methyltransferases have shown the base-flipped complexes to occupy a number of conformational states, requiring at least three exponential components (usually four) to describe the 2AP decay (16–19). In these systems, base flipping is a complex and dynamic process. In comparison, the PfoI complex is extraordinary in its conformational simplicity and in the highly unquenched environment of the flipped 2AP. Base flipping by PfoI results in the complete expulsion of the entire population of target 2AP bases from the DNA duplex. The flipped 2AP base is located in an environment that is almost uniform and where its fluorescence lifetime is largely similar to that of the free 2AP-ribonucleoside. It does not make excursions into the stacked interior of the duplex and does not interact significantly with apolar enzymatic residues, which would shorten its lifetime. This evidence, together with the lack of specificity for the flipped base, suggests that the binding pocket of PfoI is large and filled with water or that the flipped base does not reside within a binding pocket but lies on the exterior of the enzyme. The apparent lack of specific interactions between enzymatic residues and the flipped base, and the completeness of flipping, despite the very low probability (<1%) of finding the target base spontaneously extrahelical, imply base flipping by PfoI to be an active process. The enzyme must actively expel the target base from the DNA duplex rather than opportunistically catching it in a transient, extrahelical state.
Nucleotide flipping by PspGI
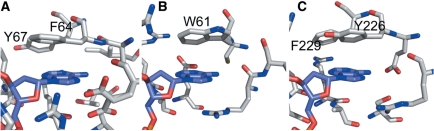
The change in decay parameters on binding of PspGI to each duplex are clearly indicative of extraction of 2AP from its highly stacked intrahelical conformation, but the environment of the flipped base differs markedly from that in the PfoI complexes. As illustrated in Figure 4A, the co-crystal structure of PspGI with its natural substrate shows that the flipped adenine lies close to two aromatic residues, F64 and Y67 in the binding pocket (14). Previously, we have found that the fluorescence of 2AP flipped into the active site of methyltransferase TaqI is efficiently quenched by such interactions with tyrosine and phenylalanine residues, resulting in subnanosecond decay times (16). We anticipate that 2AP flipped into the binding pocket of PspGI will be subject to similar quenching and the decay parameters are consistent with this expectation. The two subnanosecond decay components, which account for 90% of the population of ternary complexes, can be ascribed to quenched conformations of 2AP in the binding pocket, together with some contribution from imperfectly stacked intrahelical states. The significant difference between the decay components of the two duplexes when complexed with this enzyme (compared with the essentially identical parameters of the complexes with PfoI) supports the notion of partitioning of the target base between extrahelical and intrahelical states.
Figure 4.
The flipping pockets of PspGI (A), Ecl18kI (B) and EcoRII-C (C) (enzymes shown in grey) with adenine (blue) in the flipping pocket. Amino acids shown contain atoms that lie within 5 Å of the adenine base.
Nucleotide flipping by wt Ecl18kI and EcoRII-C
The co-crystal structure of Ecl18kI with the natural substrate (9) shows the flipped base closely stacked with tryptophan (W61) in the binding pocket, as illustrated in Figure 4B. 2AP fluorescence is known to be rapidly quenched by electron transfer from tryptophan (45). This occurs on the picoseconds timescale, at a rate comparable with (or possibly faster than) electron transfer quenching of intrahelical 2AP by guanine (43). Thus, the observation of a 70-ps decay time for 2AP in the wild-type Ecl18kI ternary complexes in the solution is entirely consistent with a base-flipped conformation equivalent to that observed for the natural substrate in the crystalline complex. Moreover, the vast majority of the population exists in this conformation. An alternative interpretation would be that the 2AP remains entirely unflipped in a tightly stacked intrahelical state. However, the X-ray structure, the change in decay parameters on enzyme binding, the similarity of the decay parameters of both duplexes when enzyme-bound (although they are different when free) and the effect of mutation on the decay parameters (vide infra) constitute convincing evidence for base flipping. A contribution from intrahelical conformations cannot be ruled out, but the close similarity between the decay parameters of the two duplexes and the lack of heterogeneity suggest that this is not significant. Hence, in the wt Ecl18kI, base flipping of 2AP is an efficient process and the binding pocket of Ecl8kI provides an environment that is similar to the DNA duplex in order to stabilize the extrahelical base.
The decay parameters of the EcoRII-C complexes are very similar to those of the wt Ecl18kI complexes, with the vast majority of the emitting population showing a very short decay time (80 ps). The crystal structure of EcoRII-C with DNA (15) shows a tyrosine residue (Y226) that coincides with the tryptophan residue (W61) in the binding pocket of Ecl18kI (Figure 4C). The oxidation potential of tyrosine (0.93 V) is slightly less than that of tryptophan (1.0 V) (46). Therefore, it can be inferred that, like Ecl18kI, EcoRII-C flips the target 2AP base into the binding pocket where it is tightly stacked with tyrosine and subject to efficient electron transfer quenching. We have not observed such rapid quenching of 2AP by tyrosine in other enzymes, such as PspGI and TaqI methyltransferase, implying that in these systems, the geometrical relationship between 2AP and tyrosine is less conducive to fast electron transfer.
Ecl18kI: the effect of mutations in the binding pocket
Changes in the structure of the binding pocket of Ecl18kI, as a result of mutation of W61, confer significant changes on the 2AP fluorescence decay. In the W61Y mutant, the replacement of the tryptophan residue in the binding pocket by tyrosine results in a substantial reduction in the efficiency of quenching of the flipped 2AP and an increase in its conformational mobility. The decay parameters are very similar to those observed for PspGI and can be interpreted in a similar fashion.
In the W16A mutant, the elimination of an interacting aromatic residue greatly reduces the quenching of the flipped 2AP, with ∼20% of the population occupying an unquenched state (with 9 ns lifetime). With this lifetime, 2AP could be in an aqueous, extrahelical state, between the DNA duplex and the binding pocket, or in a hydrated environment within the flipping pocket. The W61A mutation likely creates a void in the flipping pocket that could readily fill with water molecules. The significant increase in the value of τ2, relative to W61Y, supports the assignment of this component primarily to conformations with 2AP extrahelical, while the negligible change in τ1 suggests a substantial intrahelical contribution to this component.
There is a high degree of conformational heterogeneity in the W61A complexes, particularly, that with duplex 2/2, in which the 2AP population is rather uniformly distributed amongst the four photophysically distinct states. It is evident that the W61A mutation weakens the interaction between the enzyme and the flipped base and there is no strong energetic preference for a particular orientation of the 2AP in the binding pocket. There is a clear difference in conformational distribution between the complexes of T/2 and 2/2 with W61A, with the former showing a greater population of intrahelical states. The final distribution of the 2AP population in these complexes is thus sensitive to the strength of the base pair that is targeted for flipping. The effects of destabilization of the interactions between the binding pocket and the flipped nucleotide, in the W61A mutant, reveal that base flipping by Ecl18kI is controlled by two factors: the stability of the base pair that is the target for flipping and the interaction of the flipped base with the residues in the enzymatic flipping pocket. Hence, consistent with our previous kinetic studies (13), we find that Ecl18kI uses a two-step mechanism to achieve nucleotide flipping; first, breaking the DNA duplex and second, capturing the extruded base.
Ecl18kI mutant W61A: relationship between base flipping and cleavage activity
The greater occupancy of intrahelical states by the target base in the W61A-T/2 complex correlates with a much reduced binding affinity for T/2 and a loss of cleavage activity (13). Complexes of both T/2 and 2/2 with W61A show much greater conformational flexibility than any of the other complexes studied here and a very low cleavage rate. Thus, there appears to be a relationship between the tightness of binding of the flipped base in the enzyme pocket and the cleavage rate. The cleavage activity of the W61A mutant of Ecl18kI on a DNA duplex containing abasic sites at the target sites for flipping is near to that of the wild-type enzyme (13). Hence, base capture is not critical for catalysis to proceed. Yet, the dramatic loss of activity that results from introducing the W61A mutation seems contradictory to this argument. Thus, we propose a model for flipping where the initial disruption of the base pair is a transient, reversible event, temporarily expelling the target base(s) from the DNA duplex. Further to our previous predictions (13), the time-resolved results now show definitively that the base capture in the W61A mutant of Ecl18kI is an inefficient process. In the absence of a binding pocket that is capable of base capture, the transiently flipped base readily returns to the DNA duplex. DNA cleavage is slow relative to the rapid movement of the transiently flipped base to and from the duplex and only takes place once a stable nucleotide flip, in both strands of the duplex, and subsequent collapse of the DNA duplex is achieved.
Ecl18kI mutant W61Y: altered specificity results from unfavourable interactions with thymine in the binding pocket
The similar fluorescence decay parameters of the ternary W61Y-DNA-Ca2+ complexes are surprising given the markedly different cutting activities of the W61Y variants on the duplexes examined (∼100-fold less on the T/2 duplex as compared with the 2/2 duplex) (13). In fact, Tamulaitis et al. (13) have previously noted that this mutant shows altered specificity compared with wt Ecl18kI, preferring not to cut on duplexes containing a thymine at the flipping site, despite strong binding of these substrates. The time-resolved fluorescence results confirm that the disparity in cutting activity of the W61Y mutant on the T/2 and 2/2 duplexes is not due to different interactions of the enzyme with the 2AP bases and, therefore, is due to its treatment of the flipped thymine base. Further, the flipped 2AP is in an identical, environment for both of the complexed duplexes. Since cleavage is slow in the T/2 duplex, we infer that, despite stable 2AP flipping, the thymine base is not stably flipped by the W61Y mutant enzyme. Hence, in this complex, with the T/2 duplex, the two Ecl18kI monomeric units interact with their target bases in quite different ways. This clearly demonstrates that the two units of the dimeric Ecl18kI complex are capable of independently producing stable flips of their target nucleotides. Regarding the question of cooperativity between the monomers in the base flipping process, we cannot say whether the stable flipping of 2AP influences the probability of a thymine flip, but it is evident that the unstable flip of thymine certainly does not decrease the likelihood of a stable 2AP flip.
PspGI and EcoRII-C specificity at the flipping site
Both PspGI and EcoRII-C target the same recognition sequence, i.e. 5′-CCWGG-3′ (where W stands for A or T). The crystal structures of PspGI and EcoRII-C complexed with their cognate DNA show that both enzymes flip the central bases of this recognition sequence (14,15), as shown in Figure 4A and C.
Unlike the Ecl18kI binding pocket, in the PspGI and EcoRII-C pockets, there are several amino acids that form ‘walls’ to the pocket. These amino acids may be necessary for the recognition of the A/T bases over G/C bases. Tamulaitis et al. (13) previously proposed that a ‘double-check’ mechanism is used by PspGI and EcoRII-C during the cleavage of their substrates. In this model, the strength of the target base pair for flipping is critical for stable binding of the enzymes to the DNA. Prior to cleavage, the enzyme makes a second check of the flipped base identity once it is located within its flipping pocket. The time-resolved measurements focus solely on the second step of this check, the recognition tied to flipping, since we employ conditions that ensure complete binding of the DNA. One key question with regard to the second check, is whether this is achieved through differential affinities for bases within the binding pocket, leading to more or less flipping of the target base, or whether all bases are strongly bound in the binding pocket but that binding of non-target G/C bases occurs in such a way as to prevent catalysis from occurring.
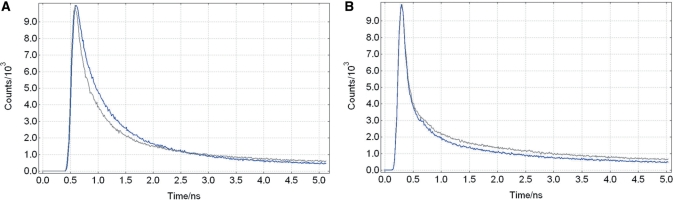
The fluorescence decays of the PspGI-T/2-Ca2+ and PspGI-2/2-Ca2+ complexes are dissimilar on the short timescale (<1 ns), especially when compared with those for the analogous EcoRII-C complexes, as shown in Tables 6 and 8 and Figure 5. There is a greater proportion of the shortest lifetime in the PspGI-2/2-Ca2+ decay, as compared with the PspGI-T/2-Ca2+ complex, which indicates a greater proportion of intrahelical 2AP in the 2/2 complex. PspGI cleaves the 2/2 duplex ∼20-fold slower than it does the T/2 duplex (13). This is consistent with the relationship between base flipping and cleavage activity that was discussed in the context of Ecl18kI (see above). This seems counter-intuitive when considering the relatively weak 2AP-2AP base pair in the 2/2 duplex, compared with the Watson–Crick-type T-2AP base pair of the T/2 duplex. It appears that the conformational distribution is determined, not by the strength of the base pair of the target bases but, rather, through the interaction of the flipped base with the PspGI. This interaction overrides the effect of the base pair strength on the overall dynamics of the system and favours more intrahelical 2AP in the 2/2 duplex than in the T/2 duplex. Since PspGI cleaves duplexes containing only A/T at the target site for flipping it is probable that discrimination against flipping of 2AP results from the presence of the 2-amino group that 2AP has in common with guanine.
Figure 5.
The first 5 ns of the fluorescence lifetime decays of PspGI (A) and EcoRII-C (B) bound in ternary complexes to the T/2 (blue) and 2/2 (grey) duplexes.
The time-resolved fluorescence results provide evidence of two important features of the second step of the double-check mechanism used by PspGI for recognition of its target base for flipping. First, 2AP is less firmly held within the PspGI binding pocket than in Ecl18kI or EcoRII-C; the target 2AP bases are flipped but only transiently, as a result of base recognition by PspGI. Second, the differential treatment of the flipped 2AP in the two complexes investigated illustrates that there is apparent communication between the two subunits of the dimeric PspGI complex. That is to say, that the stable flipping of thymine, which is a natural target base for flipping by PspGI, results in a different distribution of the 2AP population in the T/2-containing complex as compared with the transient flipping of the two 2AP bases of the 2/2 duplex. Whether this effect is the result of DNA duplex collapse, whose likelihood is increased by the stable flipping of thymine, or of some direct enzyme-enzyme contact is uncertain.
In contrast to PspGI, the catalytic subunit of EcoRII-C appears to have no mechanism by which to distinguish 2AP from A or T. The decay parameters of the EcoRII-C-T/2-Ca2+ and EcoRII-C-2/2-Ca2+ complexes are remarkably similar and in both the cases flipping and binding of the flipped 2AP are efficient. Furthermore, binding affinities and cleavage rates of the 2AP-containing duplexes are similar to those on duplexes containing the natural EcoRII-C recognition sequence (13).
SUMMARY
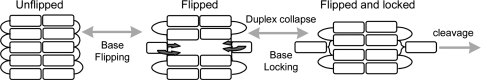
Base flipping and DNA cleavage by all of the enzymes studied go hand-in-hand. The general mechanism employed for DNA recognition and cleavage is shown schematically in Figure 6.
Figure 6.
Schematic representation of the DNA bases (white rectangles) during the flipping and locking steps that precede DNA cleavage. The target base can be unflipped (as largely seen in the Ecl18kI W61A complex), in a highly dynamic flipped (but not locked) state (predominant in the PspGI complex) or locked in the enzyme's flipping pocket (as in wt Ecl18kI and EcoRII-C complexes).
For each of the enzymes studied, their target bases must be flipped from the duplex before DNA duplex collapse occurs, which is necessary for catalysis to proceed. However, we have shown that there are significant differences in the mechanisms used by this group of enzymes to produce a base-flip that is sufficiently stable to allow the collapse of the DNA duplex. Table 9 compares the key mechanistic attributes of the enzymes.
Table 9.
A comparison of the mechanistic behaviour of the enzymes investigated
| Enzyme | Does 2AP occupy the given state in the ternary complex? |
Active flipping from duplex? | Flipping pocket capable of base capture? | Do enzymes show cooperativity to capture flipped bases? | ||
|---|---|---|---|---|---|---|
| UFa | F | FL | ||||
| PfoI | No | Yes | Yes | Possibly | Not applicable | |
| PspGI | No | Yes | Yes | Unknown | Yes | Yes |
| wt-Ecl18kI | No | No | Yes | Yes | Yes | Unlikely |
| W61Y | No | No | Yes | Yes | Yes | Unlikely |
| W61A | Yes | Yes | No | Yes | No | Not applicable |
| EcoRII-C | No | No | Yes | Unknown | Yes | Unknown |
aUF, Unflipped; F, flipped; FL, flipped and locked as illustrated in Figure 6.
We have shown that PfoI flips the base into an environment where enzymatic contacts to the base are minimal. In contrast, the other enzymes we investigated flip the base into an orientation that affords interaction with aromatic residues in the binding pocket. In Ecl18kI and EcoRII-C, the flipped 2AP is stacked particularly tightly with tryptophan or tyrosine, respectively, and sees an environment that closely resembles that within the DNA duplex and in which its fluorescence is highly quenched. Regardless of the close functional relationship between these enzymes, the flipping pocket of PfoI is quite distinct from that of the other Ecl18kI-type enzymes (i.e. Ecl18kI, EcoRII-C and PspGI).
Studies of W61 mutants of Ecl18kI have shown that base flipping is dependent on both the strength of the base pair that is the target for flipping and the ability of the enzyme to capture the flipped base. These studies have also demonstrated that base flipping is part of a concerted catalytic cycle and must be complete before DNA cleavage can proceed. Both the bases of the DNA duplex must be flipped and stably bound within the flipping pocket before cutting takes place. The linking of these processes may provide an opportunity for rational design of the base selectivity at the flipping position in the future.
PspGI discriminates between A/T and G/C bases once the bases are flipped from the duplex. Flipping of target 2AP bases is a dynamic process, the 2AP being shuttled between the DNA duplex and the enzyme's flipping pocket. The presence of the 2-amino group in 2AP, as in guanine, is a likely cause of the reduced affinity of the binding pocket for the flipped base.
EcoRII-C behaves in a similar way to wt Ecl18kI, in that base flipping of 2AP appears to be an efficient process that results in similar 2AP populations in complexes containing either the T/2 or 2/2 duplexes. Hence, EcoRII-C does not distinguish 2AP from A or T, implying that it is not sensitive to the presence of the 2-amino group.
Time-resolved fluorescence measurements of 2AP provide a means by which to examine the heterogeneity of the highly dynamic base flipping process, an insight that is not achievable using either crystallographic or ensemble kinetic measurements. However, the time-resolved measurements are used to greatest effect in combination with crystallographic, mutational and kinetic analyses and using this approach we have begun to build a remarkably detailed understanding of the structural and dynamic behaviour of the known base-flipping restriction enzymes.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Engineering and Physical Sciences Research Council (EP/C53543X/1 postdoctoral fellowship at the life-sciences interface to R.K.N.); the FP6 Marie Curie Research Training Networks grant ‘DNA Enzymes’; the Lithuanian State Science and Studies Foundation (T-14/07 and T-27/08); the European Commission (Marie Curie Intra-European Fellowship to R.K.N.]; EastChem [studentship to K.C.]. Funding for open access charge: Katholieke Universiteit Leuven.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors would like to acknowledge Dr David Dryden, Dr Patricia Richardson and Dr Mindaugas Zaremba for helpful discussions and comments on the manuscript. We thank New England Biolabs for the PspGI clone, Dr Monika Reuter (Institute of Virology, Berlin, Germany) for the EcoRII clone and Fermentas UAB for making the PfoI clone available. We are also grateful to Dr Sonata Jurenaite-Urbanaviciene for PfoI cloning.
REFERENCES
- 1.Klimasauskas S, Kumar S, Roberts RJ, Cheng XD. Hhal methyltransferase flips its target base out of the DNA helix. Cell. 1994;76:357–369. doi: 10.1016/0092-8674(94)90342-5. [DOI] [PubMed] [Google Scholar]
- 2.Reinisch KM, Chen L, Verdine GL, Lipscomb WN. The crystal-structure of HaeIII methyltransferase covalently complexed to DNA – an extrahelical cytosine and rearranged base-pairing. Cell. 1995;82:143–153. doi: 10.1016/0092-8674(95)90060-8. [DOI] [PubMed] [Google Scholar]
- 3.Goedecke K, Pignot M, Goody RS, Scheidig AJ, Weinhold E. Structure of the N6-adenine DNA methyltransferase M.TaqI in complex with DNA and a cofactor analog. Nat. Struct. Biol. 2001;8:121–125. doi: 10.1038/84104. [DOI] [PubMed] [Google Scholar]
- 4.Horton JR, Liebert K, Hattman S, Jeltsch A, Cheng XD. Transition from nonspecific to specific DNA interactions along the substrate-recognition pathway of Dam methyltransferase. Cell. 2005;121:349–361. doi: 10.1016/j.cell.2005.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yang Z, Horton JR, Zhou L, Zhang XJ, Dong AP, Zhang X, Schlagman SL, Kossykh V, Hattman S, Cheng XD. Structure of the bacteriophage T4 DNA adenine methyltransferase. Nat. Struct. Biol. 2003;10:849–855. doi: 10.1038/nsb973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carpenter MA, Bhagwat AS. DNA base flipping by both members of the PspGI restriction-modification system. Nucleic Acids Res. 2008;36:5417–5425. doi: 10.1093/nar/gkn528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hosfield DJ, Guan Y, Haas BJ, Cunningham RP, Tainer JA. Structure of the DNA repair enzyme endonuclease IV and its DNA complex: double-nucleotide flipping at abasic sites and three-metal-ion catalysis. Cell. 1999;98:397–408. doi: 10.1016/s0092-8674(00)81968-6. [DOI] [PubMed] [Google Scholar]
- 8.Banerjee A, Yang W, Karplus M, Verdine GL. Structure of a repair enzyme interrogating undamaged DNA elucidates recognition of damaged DNA. Nature. 2005;434:612–618. doi: 10.1038/nature03458. [DOI] [PubMed] [Google Scholar]
- 9.Bochtler M, Szczepanowski RH, Tamulaitis G, Grazulis S, Czapinska H, Manakova E, Siksnys V. Nucleotide flips determine the specificity of the Ecl18kl restriction endonuclease. EMBO J. 2006;25:2219–2229. doi: 10.1038/sj.emboj.7601096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Carpenter M, Divvela P, Pingoud V, Bujnicki J, Bhagwat AS. Sequence-dependent enhancement of hydrolytic deamination of cytosines in DNA by the restriction enzyme PspGI. Nucleic Acids Res. 2006;34:3762–3770. doi: 10.1093/nar/gkl545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tamulaitis G, Zaremba M, Szczepanowski RH, Bochtler M, Siksnys V. Nucleotide flipping by restriction enzymes analyzed by 2-aminopurine steady-state fluorescence. Nucleic Acids Res. 2007;35:4792–4799. doi: 10.1093/nar/gkm513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Daujotyte D, Liutkeviciute Z, Tamulaitis G, Klimasauskas S. Chemical mapping of cytosines enzymatically flipped out of the DNA helix. Nucleic Acids Res. 2008;36:e57. doi: 10.1093/nar/gkn200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tamulaitis G, Zaremba M, Szczepanowski RH, Bochtler M, Siksnys V. How PspGI, catalytic domain of EcoRII and Ecl18kI acquire specificities for different DNA targets. Nucleic Acids Res. 2008;36:6101–6108. doi: 10.1093/nar/gkn621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Szczepanowski RH, Carpenter MA, Czapinska H, Zaremba M, Tamulaitis G, Siksnys V, Bhagwat AS, Bochtler M. Central base pair flipping and discrimination by PspGI. Nucleic Acids Res. 2008;36:6109–6117. doi: 10.1093/nar/gkn622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Golovenko D, Manakova E, Tamulaitiene G, Grazulis S, Siksnys V. Structural mechanisms for the 5′-CCWGG sequence recognition by the N- and C-terminal domains of EcoRII. Nucleic Acids Res. 2009 doi: 10.1093/nar/gkp699. doi: 10.1093/nar/gkp699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lenz T, Bonnist EYM, Pljevaljcic G, Neely RK, Dryden DTF, Scheidig AJ, Jones AC, Weinhold E. 2-Aminopurine flipped into the active site of the adenine-specific DNA methyltransferase M.Taql: crystal structures and time-resolved fluorescence. J. Am. Chem. Soc. 2007;129:6240–6248. doi: 10.1021/ja069366n. [DOI] [PubMed] [Google Scholar]
- 17.Neely RK, Daujotyte D, Grazulis S, Magennis SW, Dryden DTF, Klimasauskas S, Jones AC. Time-resolved fluorescence of 2-aminopurine as a probe of base flipping in M.HhaI-DNA complexes. Nucleic Acids Res. 2005;33:6953–6960. doi: 10.1093/nar/gki995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Youngblood B, Bonnist E, Dryden DTF, Jones AC, Reich NO. Differential stabilization of reaction intermediates: specificity checkpoints for M.EcoRI revealed by transient fluorescence and fluorescence lifetime studies. Nucleic Acids Res. 2008;36:2917–2925. doi: 10.1093/nar/gkn131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hariharan C, Reha-Krantz LJ. Using 2-Aminopurine fluorescence to detect bacteriophage T4 DNA polymerase−DNA complexes that are important for primer extension and proofreading reactions. Biochemistry. 2005;44:15674–15684. doi: 10.1021/bi051462y. [DOI] [PubMed] [Google Scholar]
- 20.Tamulaitis G, Solonin AS, Siksnys V. Alternative arrangements of catalytic residues at the active sites of restriction enzymes. FEBS Lett. 2002;518:17–22. doi: 10.1016/s0014-5793(02)02621-2. [DOI] [PubMed] [Google Scholar]
- 21.Gaigalas M, Maneliene Z, Kazlauskiene R, Petrusyte M, Janulaitis A. PfoI, a unique type II restriction endonuclease that recognises the sequence 5′-T/CCNGGA-3′. Nucleic Acids Res. 2002;30:e98. doi: 10.1093/nar/gnf097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tamulaitis G, Mucke M, Siksnys V. Biochemical and mutational analysis of EcoRII functional domains reveals evolutionary links between restriction enzymes. FEBS Lett. 2006;580:1665–1671. doi: 10.1016/j.febslet.2006.02.010. [DOI] [PubMed] [Google Scholar]
- 23.Zaremba M, Sasnauskas G, Urbanke C, Siksnys V. Allosteric communication network in the tetrameric restriction endonuclease Bse634I. J. Mol. Biol. 2006;363:800–812. doi: 10.1016/j.jmb.2006.08.050. [DOI] [PubMed] [Google Scholar]
- 24.Pingoud A, Jeltsch A. Structure and function of type II restriction endonucleases. Nucleic Acids Res. 2001;29:3705–3727. doi: 10.1093/nar/29.18.3705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Allan BW, Reich NO. Targeted base stacking disruption by the EcoRI DNA methyltransferase. Biochemistry. 1996;35:14757–14762. doi: 10.1021/bi9615708. [DOI] [PubMed] [Google Scholar]
- 26.McCullough AK, Dodson ML, Scharer OD, Lloyd RS. The role of base flipping in damage recognition and catalysis by T4 endonuclease V. J. Biol. Chem. 1997;272:27210–27217. doi: 10.1074/jbc.272.43.27210. [DOI] [PubMed] [Google Scholar]
- 27.Holz B, Klimasauskas S, Serva S, Weinhold E. 2-Aminopurine as a fluorescent probe for DNA base flipping by methyltransferases. Nucleic Acids Res. 1998;26:1076–1083. doi: 10.1093/nar/26.4.1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stivers JT, Pankiewicz KW, Watanabe KA. Kinetic mechanism of damage site recognition and uracil flipping by Escherichia coli uracil DNA glycosylase. Biochemistry. 1999;38:952–963. doi: 10.1021/bi9818669. [DOI] [PubMed] [Google Scholar]
- 29.Gowher H, Jeltsch A. Molecular enzymology of the Eco RV DNA-(adenine-N6)-methyltransferase: kinetics of DNA binding and bending, kinetic mechanism and linear diffusion of the enzyme on DNA. J. Mol. Biol. 2000;303:93–110. doi: 10.1006/jmbi.2000.4127. [DOI] [PubMed] [Google Scholar]
- 30.Reddy YVR, Rao DN. Binding of EcoP15I DNA methyltransferase to DNA reveals a large structural distortion within the recognition sequence. J. Mol. Biol. 2000;298:597–610. doi: 10.1006/jmbi.2000.3673. [DOI] [PubMed] [Google Scholar]
- 31.Su TJ, Connolly BA, Darlington C, Mallin R, Dryden DTF. Unusual 2-aminopurine fluorescence from a complex of DNA and the EcoKI methyltransferase. Nucleic Acids Res. 2004;32:2223–2230. doi: 10.1093/nar/gkh531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liebert K, Hermann A, Schlickenrieder M, Jeltsch A. Stopped-flow and mutational analysis of base flipping by the Escherichia coli Dam DNA-(adenine-N6)-methyltransferase. J. Mol. Biol. 2004;341:443–454. doi: 10.1016/j.jmb.2004.05.033. [DOI] [PubMed] [Google Scholar]
- 33.Szegedi SS, Reich NO, Gumport RI. Substrate binding in vitro and kinetics of RsrI [N6-adenine] DNA methyltransferase. Nucleic Acids Res. 2000;28:3962–3971. doi: 10.1093/nar/28.20.3962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Malygin EG, Evdokimov AA, Zinoviev VV, Ovechkina LG, Lindstrom WM, Reich NO, Schlagman SL, Hattman S. A dual role for substrate S-adenosyl-L-methionine in the methylation reaction with bacteriophage T4 Dam DNA-[N6-adenine]-methyltransferase. Nucleic Acids Res. 2001;29:2361–2369. doi: 10.1093/nar/29.11.2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Christine KS, MacFarlane AW, IV, Yang K, Stanley RJ. Cyclobutylpyrimidine Dimer Base Flipping by DNA Photolyase. J. Biol. Chem. 2002;277:38339–38344. doi: 10.1074/jbc.M206531200. [DOI] [PubMed] [Google Scholar]
- 36.Malta E, Moolenaar GF, Goosen N. Base flipping in nucleotide excision repair. J. Biol. Chem. 2006;281:2184–2194. doi: 10.1074/jbc.M508901200. [DOI] [PubMed] [Google Scholar]
- 37.Walker RK, McCullough AK, Lloyd RS. Uncoupling of nucleotide flipping and DNA bending by the T4 pyrimidine dimer DNA glycosylase. Biochemistry. 2006;45:14192–14200. doi: 10.1021/bi060802s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rachofsky EL, Seibert E, Stivers JT, Osman R, Ross JBA. Conformation and dynamics of abasic sites in DNA investigated by time-resolved fluorescence of 2-aminopurine. Biochemistry. 2001;40:957–967. doi: 10.1021/bi001665g. [DOI] [PubMed] [Google Scholar]
- 39.O'Neill MA, Dohno C, Barton JK. Direct chemical evidence for charge transfer between photoexcited 2-aminopurine and guanine in duplex DNA. J. Am. Chem. Soc. 2004;126:1316–1317. doi: 10.1021/ja037802p. [DOI] [PubMed] [Google Scholar]
- 40.Kelley SO, Barton JK. Electron transfer between bases in double helical DNA. Science. 1999;283:375–381. doi: 10.1126/science.283.5400.375. [DOI] [PubMed] [Google Scholar]
- 41.Rachofsky EL, Osman R, Ross JBA. Probing structure and dynamics of DNA with 2-aminopurine: effects of local environment on fluorescence. Biochemistry. 2001;40:946–956. doi: 10.1021/bi001664o. [DOI] [PubMed] [Google Scholar]
- 42.Neely RK, Magennis SW, Dryden DTF, Jones AC. Evidence of tautomerism in 2-aminopurine from fluorescence lifetime measurements. J. Phys. Chem. B. 2004;108:17606–17610. [Google Scholar]
- 43.Fiebig T, Wan CZ, Zewail AH. Femtosecond charge transfer dynamics of a modified DNA base: 2- aminopurine in complexes with nucleotides. ChemPhysChem. 2002;3:781–788. doi: 10.1002/1439-7641(20020916)3:9<781::AID-CPHC781>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 44.O'Neill MA, Becker HC, Wan CZ, Barton JK, Zewail AH. Ultrafast dynamics in DNA-mediated electron transfer: Base gating and the role of temperature. Angew. Chem. Int. Ed. 2003;42:5896–5900. doi: 10.1002/anie.200352831. [DOI] [PubMed] [Google Scholar]
- 45.Xia TB, Becker HC, Wan CZ, Frankel A, Roberts RW, Zewail AH. The RNA-protein complex: direct probing of the interfacial recognition dynamics and its correlation with biological functions. Proc. Natl Acad. Sci. USA. 2003;100:8119–8123. doi: 10.1073/pnas.1433099100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Harriman A. Further comments on the redox potentials of tryptophan and tyrosine. J. Phys. Chem. 1987;91:6102–6104. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.