Abstract
To characterize the organization of mtDNA–protein complexes (known as nucleoids) in vivo, we have probed the mtDNA surface exposure using site-specific DNA methyltransferases targeted to the mitochondria. We have observed that DNA methyltransferases have different accessibility to different sites on the mtDNA based on the levels of protein occupancy. We focused our studies on selected regions of mtDNA that are believed to be major regulatory regions involved in transcription and replication. The transcription termination region (TERM) within the tRNALeu(UUR) gene was consistently and strongly protected from methylation, suggesting frequent and high affinity binding of mitochondrial transcription termination factor 1 (mTERF1) to the site. Protection from methylation was also observed in other regions of the mtDNA, including the light and heavy strand promoters (LSP, HSP) and the origin of replication of the light strand (OL). Manipulations aiming at increasing or decreasing the levels of the mitochondrial transcription factor A (TFAM) led to decreased in vivo methylation, whereas manipulations that stimulated mtDNA replication led to increased methylation. We also analyzed the effect of ATAD3 and oxidative stress in mtDNA exposure. Our data provide a map of human mtDNA accessibility and demonstrate that nucleoids are dynamically associated with proteins.
INTRODUCTION
The human mitochondrial genome is organized in DNA–protein complexes known as nucleoids. A variety of processes including replication, transcription and repair occur in the context of these structures (1,2). Each human nucleoid is believed to contain two to eight mtDNA molecules (3). Nucleoids are actively distributed during mitochondrial fusion and fission ensuring proper segregation of mitochondrial genomes (4). Biochemical isolation procedures have identified several nucleoid components such as DNA polymerase gamma catalytic subunit (POLGA), a putative mtDNA helicase (Twinkle), mitochondrial transcription factor A (TFAM) and mitochondrial single stranded binding protein (mtSSB) (5). While some components of the mitochondrial nucleoids have been identified, nucleoid structural organization and dynamics remain poorly understood in mammalian cells. It has been speculated that during mtDNA replication and under certain physiological conditions nucleoid composition and structure varies.
The high mobility group (HMG) family protein, TFAM, has been reported as the most abundant nucleoid component (6). Members of HMG proteins are able to bind, bend and unwind DNA (7). TFAM is involved in mtDNA maintenance, transcription, replication, and is thought to play a major role in mtDNA packaging and compaction similar to histone proteins and bacterial HU protein (2,6). Ablation of TFAM in mice results in embryonic lethality due to severe mtDNA depletion (8). Over-expression of TFAM is sufficient to increase mtDNA transcription; however, high levels of TFAM are not necessarily correlated with increased mtDNA copy number (9,10). TFAM binding to DNA has a low degree of specificity; however, it has been shown to bind with higher affinity to the light- and heavy-strand promoters, LSP and HSP (11). The ratio of TFAM/mtDNA is currently a controversial issue. In HeLa cells, 900–1700 molecules of TFAM were reported to be associated with one molecule of mtDNA (2). It has been estimated that TFAM occupies ∼25 bp, therefore 900 molecules of TFAM would be enough to coat the entire mitochondrial genome (11–13). Accordingly, atomic force microscopy experiments have shown that TFAM alone has the capacity to fully compact mtDNA (14). However, it is still not known whether mtDNA is fully saturated with TFAM in vivo. In contrast, more recently, TFAM was reported to be present at lower quantities in HeLa cells ranging from 35 to 50 molecules per mtDNA (15,16). This estimate would restrict the number of TFAM binding sites making the mtDNA less packaged. That relatively low TFAM:mtDNA ratio was demonstrated to be enough to stimulate mtDNA transcription, whereas much higher concentrations of TFAM had an inhibitory effect on transcription (15).
In the present study, we adopted an approach previously used for nuclear DNA to identify potential mtDNA–protein interactions in living cells. The use of prokaryotic DNA methyltransferases has been shown to be a powerful strategy in mapping in vivo DNA binding sites of chromatin proteins (17–19). DNA regions coated by proteins are less accessible to methyltransferases, whereas ‘naked’ DNA is easily methylated. Similar to chromatin structures, mitochondrial nucleoids are also dynamic and regulated by remodeling protein complexes. Therefore, in order to map methyltransferase accessibility to mtDNA, we targeted two bacterial DNA methyltransferases to mitochondria, M.ApaI which recognizes low frequency GGGCCC sites and M.CviPI which recognizes higher frequency GpC dinucleotide sites.
Using this approach, we identified changes in methyltransferases accessibility to the mtDNA in response to different cell treatments that modify the nucleoid environment.
MATERIALS AND METHODS
Construction of DNA methyltransferases genes targeted to the mitochondria
The gene encoding for ApaI methyltransferase (M.ApaI) was PCR amplified from the Acetobacter pasteurianus genome. pSH1052 plasmid containing an insert encoding for M.CvipI was kindly provided by Dr Michael P. Kladder, University of Florida. A mitochondrial target sequence from the cytochrome oxidase subunit VIII was added in frame to the 5′-end of the genes, and an HA tag was included at the 3′-end. The final constructs were cloned in a Mifepristone (RU486) inducible vector derived from the Gene Switch System (Invitrogen) to tightly control the expression of the genes. The gene encoding for TFAM was PCR amplified from a human cDNA library and cloned into a vector driven by a CMV promoter containing a pIRES GFP. The Mito-M.CvipI-HA gene was also cloned in a pIRES-GFP lentivirus vector under control a CMV promoter. Lentivirus was prepared by the Viral Vector core Facility Miami Project to Cure Paralysis, University of Miami.
Cell cultures and DNA transfections
Human osteosarcoma 143B, human embryonal kidney (HEK) 293T and HeLa cells were cultured in Dulbecco’s medium supplemented with 1 mM sodium pyruvate, 50 µg/ml uridine and 10% fetal bovine serum. For plasmid transfections, cells were plated at ∼70% confluence in 6-well tissue culture plates and plasmids were transfected using Fugene reagent (Roche) as described by the manufacturer. After 48 h, transfected cells with mito-M.CviPI were selected using 100 mg/ml hygromycin B (Invitrogen). Stable clones were isolated after 2 wks of selection. For Lentiviral infections, 1 × 105 cells were seeded in 6-well plates and transduced with 6 × 106 pg of viral particles.
siRNAs for TFAM, mTERF and ATAD3 knockdown
TFAM, mTERF and ATAD3 siRNAs were synthesized using the Silencer siRNA construction kit (Ambion). To knockdown TFAM, two siRNAs against exon 4 were used: AAGTTGTCCAAAGAAACCTGT and AAGATGCTTATAGGGCGGAGT corresponding to mRNA nt 273–293 and nt 431–451, respectively (10). mTERF1 was target using AAGCGGGUGAAAGCUAACAUU corresponding to mRNA nt 585–605 (20). To knockdown ATAD3 expression, cells were transfected with siRNA against 5′-UCAAUGAGGAGAAUUUACGGAAGCAAG-3′ (21). Cells were transfected with 20 nm of siRNA using Lipofectamine RNAiMAX (Invitrogen).
Immunocytochemistry
Cells were gown on coverslips and induced with 10 nM RU486 (Invitrogen). After 24 h, cells were incubated with 200 nM CMX-ROS MitotrackerTM (Invitrogen) for 20 min. Cells were washed with PBS and fixed with 4% paraformaldehyde and permeabilized with cold methanol for 5 min and stained with rat anti-HA antibody (Roche) and anti-rat IgG Alexa-fluor 488 secondary antibody (Invitrogen). Images were obtained using a confocal Axiovert 100 M microscope with a LSM510 scanning module (Carl Zeiss).
Southern blotting
Approximately 0.5 µg of genomic DNA was subjected to ApaI digestion for 4 h. Samples were resolved in a 0.8% agarose gel and transferred to a Zeta-Probe GT membrane (Bio-Rad, Hercules, CA). Membranes were hybridized with [α-32P] dCTP-labeled random primed probes generated against a 16 kb mtDNA template. Images were recorded using a Cyclone Storage Phosphor System and analyzed using the associated Optiquant software (Perkin Elmer).
Western blotting
Thirty micrograms of proteins from cell homogenates were subjected to electrophoresis in a 4–20% Tris–HCl polyacrylamide gel (Bio-Rad) and transferred to a PVDF membrane (Bio-Rad). Membranes were probed with anti-TFAM and anti-b-tubulin antibodies (Sigma, Saint Louis, MO). HRP-conjugated secondary antibodies (Sigma) were used and reactions were developed using Super Signal West Pico Chemiluminescent Substrate (Pierce, Rockford, IL).
Real-time PCR
mRNA from cells transfected with TFAM and ATAD3 siRNAs were harvested using an RNeasy kit (Qiagen). cDNAs were synthesized using Superscript reverse transcriptase (Invitrogen). Real-time PCR using CYBR green (Qiagen) was performed in order to detect TFAM and ATAD3 mRNA expression levels. Gene expression levels were normalized against beta actin values. Primer design was aided by Biosearch Technologies at http://www.biosearchtech.com/products/probe_design.asp.
Bisulfite genomic PCR
Genomic DNA was extracted and bisulfite treated with the EpiTect Bisulfite Kit (Qiagen). Bisulfite-treated DNA was PCR amplified with various primers flanking mtDNA regions of interest. Cycling conditions were: 3 min at 94°C, 45 cycles of 30 s at 94°C, 30 s at 50°C, 30 s at 72°C followed by 4 min at 72°C. PCR products were purified by GenElute PCR Clean-Up Kit (Sigma-Aldrich) and sequenced by a commercial facility (Genewiz).
Methylation quantification
Mutation Surveyor software (Softgenetics) was used to analyze the ab1 sequencing files from bisulfite converted samples. Mutation surveyor detected methylation as point mutation (GT > GC) by comparing the sequencing trace data of methylated samples to a reference (unmethylated) sample. Trace intensities were obtained using the Output Trace Data function and exported to an Excel file. The trace data from methylated samples were normalized by the trace data from unmethylated control samples. The intensity ratio of the Thymine (T) peaks corresponding to the GpC sites of methylated and control samples was calculated. The average of the intensity ratio of 8 T peaks surrounding the GpC site was also determined. The thymine peak corresponding to a methylated GpC site subtracted and divided by the average of the thymine peak values is equivalent to the level of methylation at a particular site. A selected mtDNA segment in the D-loop region was amplified, treated and analyzed independently in triplicate. This analysis showed essentially identical levels of protection (not shown).
RESULTS
Expression of DNA methyltransferases targeted to mitochondria
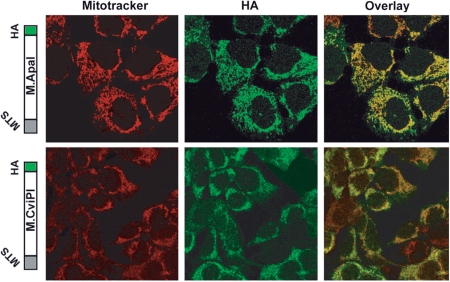
We constructed two genes encoding bacterial methyltransferases, M.ApaI and M.CviPI, to have their products targeted to the mitochondria. The M.ApaI recognizes specifically the GGGCCC sequence and it was selected because it can probe the occupancy of mTERF1 in the TERM region of the tRNALeu(UUR) gene in cells that harbor the MELAS (Mitochondrial Encephalomyopathy Lactic Acidosis and Stroke-like episodes) A3243G mutation. The M.CviPI methyltransferases GpC sites, which occur at a high frequency in the mtDNA (710 sites), was used for a more detailed in vivo mapping of methyltransferases accessibility to the mitochondrial genome. For mitochondrial targeting and immuno-detection, the COX8 mitochondrial targeting sequence (MTS) was added to the N-terminus of each gene and an HA immuno-tag was added to the C-terminus (Figure 1). In order to better control the expression levels of the mito-methyltransferases, the final constructs were cloned in a RU486 (mifepristone) inducible vector derived from the Gene Switch system (Invitrogen). The mito-M.ApaI-HA construct was stably transfected into 143B cybrids homoplasmic for the MELAS mutation and mito-M.CviPI-HA was stably transfected into HEK293T cells. Confocal microscopy confirmed the expression of both methyltransferases in the mitochondria of both cell types after 24 h induction (Figure 1).
Figure 1.
Construction and expression of methyltransferases targeted to the mitochondria. A DNA fragment coding for a mitochondria targeting sequence (MTS) derived from the COX VIII gene was added to the 5′-end of the DNA methyltransferase genes and a HA tag was added to the 3′-end of the genes. The upper panel shows co-localization of ApaI methyltransferase and mitotracker in human osteosarcoma cells transfected with mito-M.ApaI-HA. The lower panel shows a similar experiment performed in HEK293T cells transfected with mito-M.CviPI-HA.
mTERF1 binding site is strongly protected against methylation
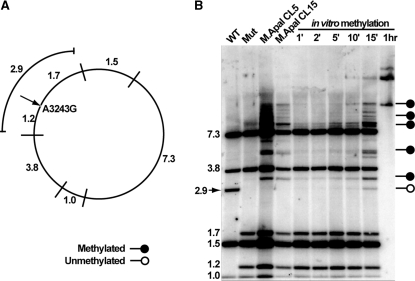
Southern blot analysis with a methylation-sensitive restriction enzyme was performed to evaluate the ability of cells stably expressing mito-M.ApaI-HA to methylate mtDNA. In this assay, total genomic DNA was digested with the cognate M.ApaI restriction endonuclease, ApaI, which recognizes the same sites as M.ApaI. Methylation of cytosine on both DNA strands by M.ApaI inhibits cleavage by ApaI restriction endonuclease. Genomic DNA extracted from wild-type, MELAS cybrids and two mito-M.CviPI-HA clones induced with mifepristone for 24 h were digested with ApaI. As an experimental control, we performed an in vitro methylation assay by treating genomic DNA with a commercial GGCC methyltransferease (M.HaeIII) for different time points followed by digestion with ApaI (Figure 2b, lanes 5–10). In both cases probes generated against the entire mtDNA were used to visualize mtDNA fragments.
Figure 2.
Southern blot methylation assay showing partial methylation of cells expressing ApaI methyltransferase. Panel A shows the expected band sizes after ApaI restriction endonuclease digestion of the human mtDNA harboring the MELAS A3243G mutation. The arrow shows the mutated 3243 site (an ApaI site), within the binding site of mTERF1. If this site is not digested, the 1.2 and 1.7 kb bands merge into a 2.9 kb band. Panel B shows the Southern blot of DNA from the mutant cells after expression of M.ApaI and controls. The absence of the 2.9 kb band in the M.ApaI expressing clones indicates methylation protection at the 3243 G position. The arrow in panel B indicates wild-type band which is absent from samples that have the MELAS mutation. Methylation of naked DNA (in vitro) does not protect the 3243G.
The Southern blot methylation assay revealed that mtDNA from wild-type and mutant control cells not transfected with mito-M.ApaI-HA were completely digested by ApaI restriction endonuclease. In contrast, mito-M.ApaI-HA cells showed partial digestion, reflecting partial methylation of ApaI recognition sites (Figure 2). The partial digestion is seen as extra bands not present in the wild-type and mutant control samples. The same pattern of partial digestion was also seen in the ‘short-incubation’ in vitro methylation assay samples confirming the partial methylation in vivo. Interestingly, the five common ApaI sites that are present in both wild-type and mutant mtDNA were partially methylated in the mito-M.ApaI-HA expressing cells, but the unique ApaI site within the mTERF1 binding site created by the MELAS mutation was completely digested indicating it was not methylated. This ApaI site generates two new bands (1.7 and 1.2 kbp) that are equivalent to cleavage of the 2.9 kbp in the wild-type mtDNA. Undigested 2.9 kbp was present in the in vitro methylation samples suggesting that M.ApaI was not able to access the ApaI mtDNA site within the mTERF binding site in living cells. As it was observed before, in the nuclear genome, we believed that protection from methylation is due to protein–DNA contacts that inhibit the access of DNA methyltransferases. The mitochondrial transcription termination factor 1 (mTERF1) is the only protein known to bind to the mtDNA region that includes this particular ApaI site.
Identification of mtDNA protected sites with a GpC methyltransferase targeted to the mitochondria
Because the M.CviPI methyltransferase recognizes all 710 GpC sites in mtDNA, we used this construct for a more detailed mapping of selected regions. Because of the larger number of sites, we performed bisulfite genomic PCR followed by quantitative electropherogram analysis rather than Southern blotting in order to determine the methylation status of mtDNA in HEK293T cells expressing mito-M.CviPI-HA. This technique is based on the deamination of cytosine, but not 5-methyl-cytosine, to uracil by sodium bisulfite (22). Cells transfected with mito-M.CviPI-HA were induced with mifepristone for 24 h followed by genomic DNA extraction and bisulfite genomic PCR. We investigated three major regulatory regions of the mtDNA: the D-loop or major non-coding region (NCR), the binding site for mTERF1 (TERM) and the origin of the L-strand replication (OL).
The height of the electropherogram peaks does not reflect the percentage of the specific base precisely. To more accurately quantify the methylation levels at each GpC site from sequencing electropherograms, we used a novel mutation quantifier function developed for the Mutation Surveyor software (Softgenetics, http://www.softgenetics.com/mutationSurveyor.html). Mutation surveyor software detects GpC methylation in ABI trace files from bisulfite treated DNA as a GT—>GC mutation compared to a reference (unmethylated) trace file. DNA methylation quantification is determined by quantifying the drop of the T peak intensity of the methylated sample compared to the unmethylated sample taking into account context dependent variations in T peak heights. We have also obtained similar results by using a recently described method known as Mquant (23). Mquant uses a similar algorithm method based on the peak area determination from sequencing electropherograms and has been shown to give results consistent with the well-established combined bisulfite restriction assay (COBRA) (24). Using the Mutation Surveyor software, we observed differential accessibility to the M.CviPI methyltransferase (Figures 3–5). The most protected GpC sites identified were located upstream the mt3L, consensus binding site for nuclear protein factors (25), region (GpC site 16536), in the LSP promoter (GpC site 418), in the HSP and nearby regions (GpC sites 418, 602, 698 and 702), The TERM (GpC sites 3240, 3245 and 3256) and in the OL region (GpC sites 5624, 5755, 5764 and 5798). Among these sites, the TERM region was completely protected from methylation (Figure 5 and Supplementary Figure S1) as observed with the ApaI methyltransferase. We observed low levels of protection from methylation in the LSP and HSP regions corresponding to the canonical TFAM binding sites (Figure 4). This result suggests that proteins occasionally coat the promoter region, but to a lesser frequency or affinity when compared to the mTERF1 binding to the TERM site. It is possible that D-loop accessibility is modulated during transcription and replication, when factors are required to access the OH, LSP or HSP. The protection at the non-coding region of the OL also suggests high affinity binding of factors to that region.
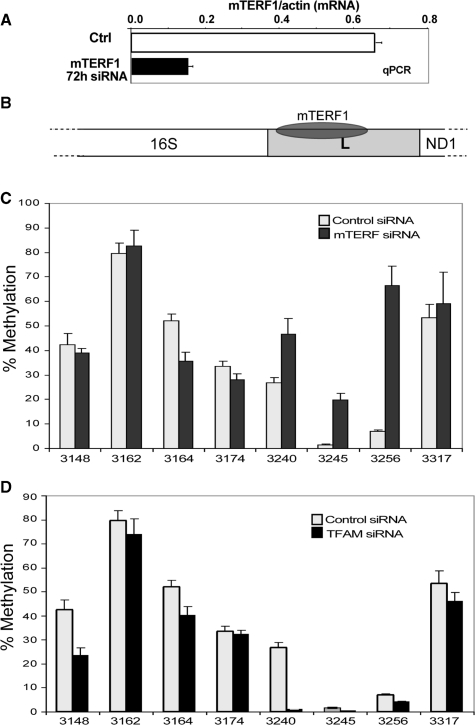
Figure 3.
TERM site in the tRNA leucine gene is strongly protected from methylation. (A) qRT-PCR showing the relative decrease in the levels of mTERF1 mRNA after 72 h of siRNA treatment. (B) Diagram shows mTERF1 binding site in the mtDNA TERM region, within the tRNALeu(UUR) (L) gene. (C) Comparison of mtDNA methylation in control and mTERF1 siRNA transfected cells. The mTERF1 siRNA-treated sample showed an increase in methylation at the GpC sites within positions 3240–3256, which correspond to the mTERF1 binding region. (D) mtDNA methylation when TFAM is silenced does not increase methylation in the TERM region.
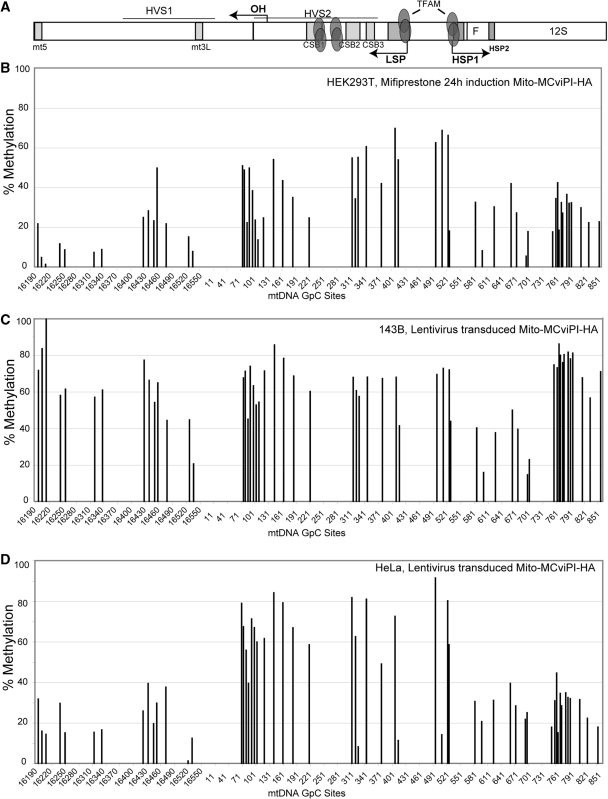
Figure 4.
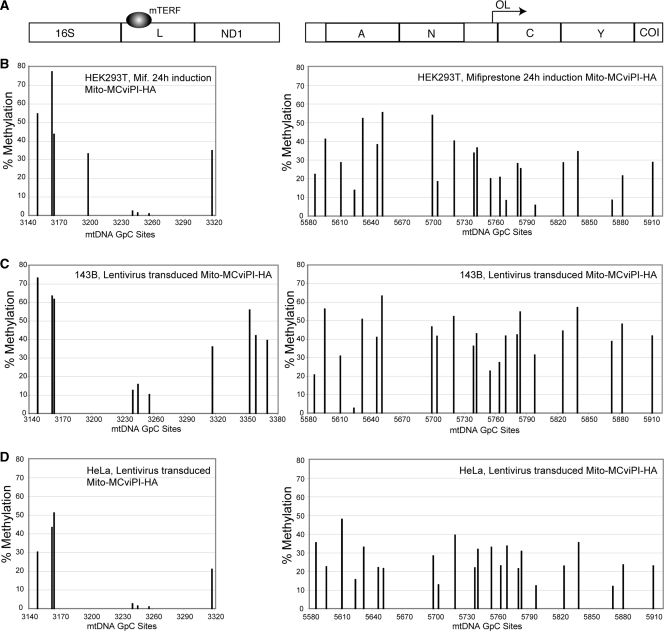
In vivo mtDNA methylation in the NCR (D-loop region) in different cell lines. Following in vivo methylation by M.CviPI-HA, either by mifiprestone induction (B) or by transduction with a lentiviral vector (C and D), total DNA was purified and bisulfite treated. Sequencing electropherograms from treated DNA were analyzed and compared to a non-methylated reference sequence. DNA methylation was quantified by the percentage drop of Ts (converted Cs) relative to its neighboring peaks (see methods). (A) Diagram showing a map of the major non-coding region (NCR) of the mtDNA indicating the position of conserved sequence boxes (CSBs), promoter regions (LSP and HSP) and other conserved cis-elements. It also shows high affinity TFAM binding sites. (B–D) mtDNA methylation levels in HEK293T (B), 143B (C) and HeLa (D) cells.
Figure 5.
In vivo mtDNA methylation in the TERM and OL regions of the mtDNA in different cell lines. (A) Map of the regions of the mtDNA comprising the TERM site and OL. (B–D) mtDNA methylation quantification in HEK293T (B), 143B (C) and HeLa (D) cells was determined as in legend to Figure 4.
To confirm that the lack of methylation seen in the mTERF1 binding site in MELAS cells was due to mTERF1 occupancy (Figure 3B), we silenced the expression of mTERF1 using RNA interference. mTERF1 specific siRNA was transfected into HEK293T stably transfected with mito-M.CvipI-HA gene. Real-time PCR revealed a decrease in mTERF1 mRNA of 75% compared to control at 72 h post siRNA transfection (Figure 3A). As expected, down-regulation of mTERF1 resulted in increased mtDNA methylation in the TERM region (Figure 3C). In contrast to the increased methylation following down-regulation of mTERF1, down-regulation of TFAM did not affect methylation in the TERM region (Figure 3D).
Comparison of the mtDNA methylation pattern in different cells lines
Because there is a discrepancy regarding the ratio of TFAM/mtDNA in different cell types, it is possible that TFAM may occupy all available mtDNA sites in some cells, making it more compact and less accessible to the methyltransferase. To investigate whether methyltransferase accessibility to mtDNA might be cell-type specific, we transduced 143B and HeLa cells with a lentivirus vector encoding for the Mito-M.CviPI-HA gene, allowing a direct comparison with the results from our inducible mito-M.CviPI-HA HEK293T model. Both transduced and induced cells shared some methylation protection sites, indicating a conserved structural organization and protein–DNA interactions at particular sites (Figures 4 and 5). Similar pattern of protection was observed at the mt3L, OH, LSP, HSP1, HSP2 and TERM regions. However, 143B and HeLa showed lower levels of methylation in the 12S rRNA (e.g. nts 761–851) and D-loop upstream of HVS1 (e.g. 16 190–16 340). The TFAM binding site at GpC site 418 (LSP) showed the highest degree of protection in HeLa cells when compared to 143B and HEK293T. We also observed strong protection at positions 330 (CSB2) and 514 (upstream of HSP1) in HeLa cells. Conversely, all other sites in the promoter, OH region and vicinity showed a high degree of methylation in HeLa cells. Because we observed a more exposed promoter/OH region and less exposed TFAM binding sites immediately upstream of the promoters, it is possible that HeLa mtDNA has higher rates of transcription when compared to mtDNA from the other cells. VDAC and cytochrome c oxidase subunit II (COXII) levels were comparable in all three cell lines, whereas TFAM and mtDNA levels were lower in HeLa than in 143B and HEK293T cells (Supplementary Figure S3). This suggests that HeLa cells have a higher relative proportion of mtDNA undergoing transcription at any given time and therefore, more frequent binding of TFAM to the promoter region.
The mtDNA nucleoids have a less compact state during mtDNA replication after EtBr treatment
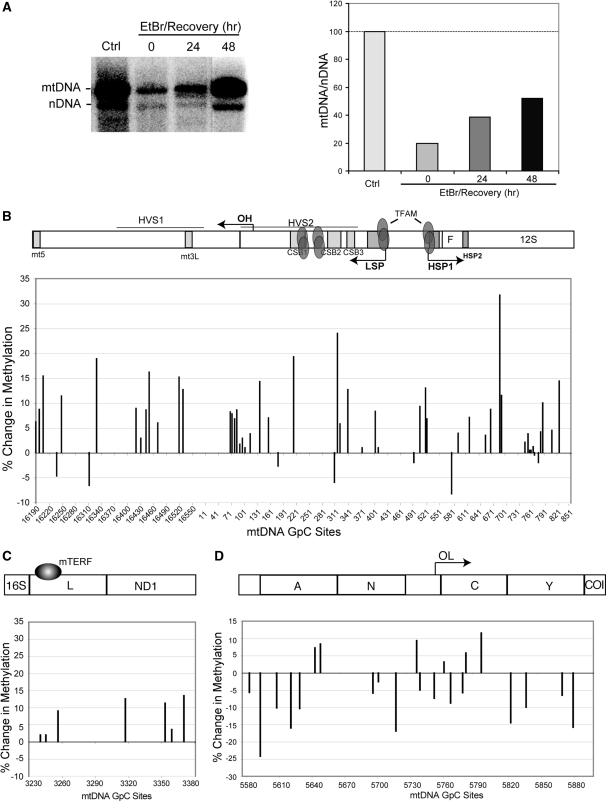
We next used mito-M.CviPI-HA as a tool to analyze mtDNA nucleoid remodeling during mtDNA replication. It is not known whether the mammalian mtDNA undergoes any form of remodeling; however, it has been show that yeast nucleoids are remodeled in response to metabolic cues. Yeast mitochondrial nucleoids have a more open structure under aerobic growth conditions and a more compact structure under glucose repression (26). Mammalian mtDNA nucleoids are also dynamic structures and likely subjected to remodeling. To test this hypothesis, we analyzed mito-M.CviPI-HA accessibility to mtDNA undergoing active replication following transient mtDNA depletion. mito-M.CviPI-HA cells were treated with 50 ng/ml EtBr for 10 days, after which the drug was removed. Southern blots confirmed that the mtDNA: nDNA ratios changed in a predicted manner (Figure 6A). We determined the methylation pattern of samples treated with EtBr for 10 days and collected 48 h after EtBr treatment was removed and mtDNA levels were recovering (48 h EtBr/Rec). With exception of the OL region (Figure 6D), an increase in methylation levels was observed in the 48 hr EtBr/Rec sample (Figure 6B–C). These results suggest that the mtDNA nucleoids form a more open structure during active mtDNA replication.
Figure 6.
mtDNA methylation levels in cells treated with EtBr followed by mtDNA repopulation after EtBr withdraw. (A) Southern blot and quantification of mtDNA relative levels from cells subjected to transient EtBr treatment. Cells were allowed to grow for 24 or 48 h without the drug to allow for active mtDNA repopulation. Southern was probed with an mtDNA and a nuclear 18S rDNA probe. (B–D) Relative changes (to the untreated control) in methylation of selected mtDNA regions from cell subjected to EtBr + 48 h recovery treatment.
mtDNA becomes less accessible when TFAM levels are either up- or down-regulated
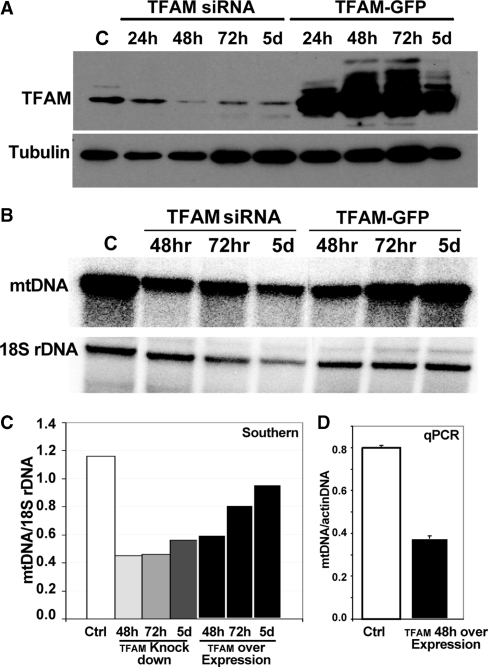
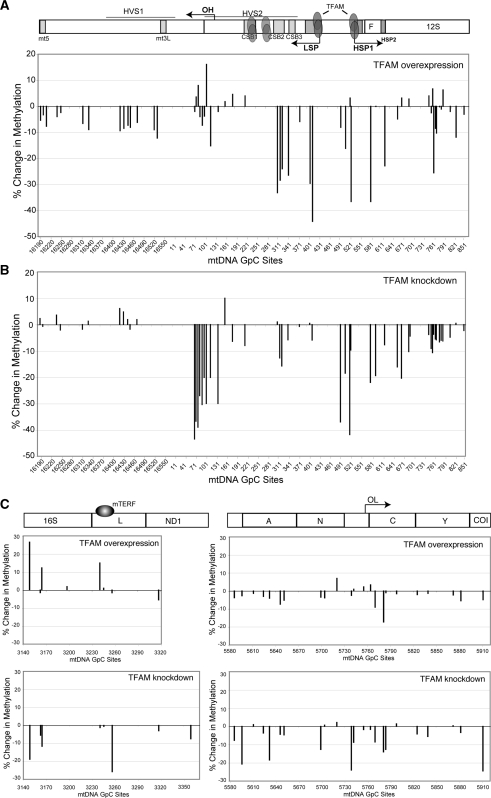
To test whether alterations in TFAM levels would affect the nucleoid structure, we analyzed mtDNA methylation when TFAM was overexpressed and when its levels were reduced. In order to overexpress TFAM, HEK293T cells expressing an inducible mito-m.M.CviPI-HA were transiently transfected with a pIRES-GFP vector encoding TFAM. Transfected cells were induced with mifepristone for 24 h for methyltransferase expression and GFP positive cells were sorted by flow-cytometry. Western blotting of GFP positive cells confirmed TFAM expression was strongly up-regulated 72 h after transfection (Figure 7A). Most of the GpC sites showed a slight decrease in methylation when compared to controls (Figure 8, upper half of panels). However, the decrease in methylation was more prominent in the mtDNA promoter region, where TFAM is known to bind with higher affinity (Figure 8A, upper half). In contrast, we performed a TFAM knockdown experiment in order to determine whether TFAM depletion would increase mito-M.CviPI-HA accessibility (Figure 8, lower half of panels). Western blotting confirmed that TFAM protein levels were decreased 48 h after siRNA transfection (Figure 7A). We were surprised to observe a decrease in mtDNA methylation in cells treated with TFAM siRNA after 72 h compared to control cells (Figure 8, lower half of panels). DNA analyses were performed to test whether TFAM depletion correlated with a decrease in mtDNA levels. Indeed, mtDNA levels were depleted by ∼35% compared to controls (Figure 7B and C). mtDNA was also decreased when TFAM was overexpressed (Figure 7C and D).
Figure 7.
Manipulating TFAM levels. (A) Western blot showing TFAM down-regulation by siRNA and up-regulation by transfection with a plasmid encoding for TFAM. Tubulin was used as a loading control. (B and C) Southern blot showing mtDNA depletion in cells where TFAM is up- or down-regulated. (D) qPCR of mtDNA levels, expressed as a ratio to the actin gene amplification, when TFAM is overexpressed.
Figure 8.
Changes in mtDNA methylation when TFAM levels are altered. Relative changes in methylation were determined in the following selected mtDNA regions: NCR (A), TERM (B) and OL (C). Changes were determined when TFAM was either overexpressed (upper part of panels) or down-regulated (lower part of panels). Methylation of mtDNA was determined as described in legend to Figure 4 and expressed as changes from the control.
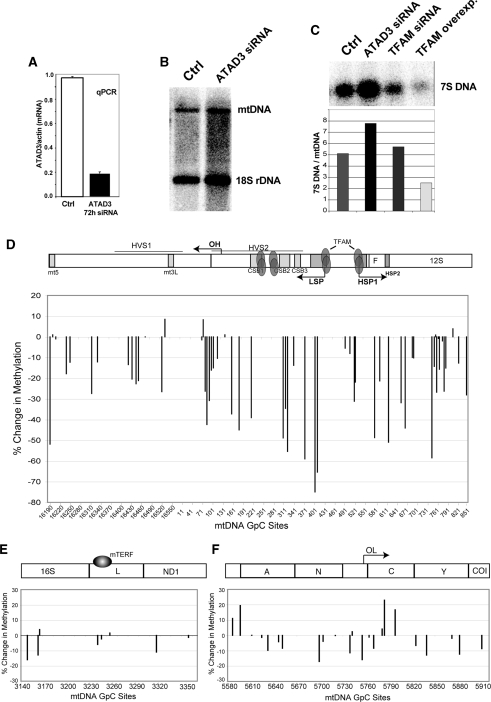
mtDNA methylation is decreased after ATAD3 knockdown
We decided to investigate whether another nucleoid protein, besides TFAM, could also affect mtDNA structure and nucleoid organization. It was recently reported that gene silencing of ATAD3, a mitochondrial membrane AAA + protein, alters the organization of mtDNA nucleoids, even though it does not seem to interact directly with mtDNA (27). Previous work showed that knockdown of ATAD3 resulted in loss of mtDNA staining by PicoGreen, suggesting a change in the mtDNA conformation and topology that excludes PicoGreen intercalation (21). In order to determine whether changes in mtDNA topology influences mito-M.CviPI-HA accessibility, we knocked down ATAD3 using siRNA. Depletion of ATAD3 levels was confirmed by real time PCR (Figure 9A). As previously described (21), a reduction in ATAD3 levels was not accompanied by mtDNA depletion (Figure 9B). However, we observe a 1.5 increase in the levels of 7S DNA (Figure 9C). Decreased mtDNA methylation was observed at most GpC sites, suggesting that change in mtDNA topology does reduce mito-M.CviPI-HA accessibility (Figure 9D–F). Since M.CviPI shows considerable preference for supercoiled plasmid in vitro (Supplementary Figure S2), we speculate that knockdown of ATAD3 results in opening of the mtDNA super-coiled structure.
Figure 9.
ATAD3 knockdown resulted in decreased mtDNA methylation. (A) Quantitative RT-PCR of a HEK293T treated with ATAD3 siRNA showed decreased levels of ATAD3 mRNA. (B) Southern blot of treated cells showed that mtDNA levels in cells transfected with ATAD3 siRNA were not altered. (C) Southern blot of siRNA for ATAD3 treated cells detected an increase in 7S DNA/mtDNA relative levels. Down-regulation of TFAM did not affect the ratio of 7S DNA/mtDNA whereas up-regulation of TFAM caused a decrease in 7S DNA/mtDNA relative levels. (D–F) Quantification of relative changes in methylation in cells transfected with ATAD3 siRNA in the following selected regions: NCR (D), TERM (E) and OL (F).
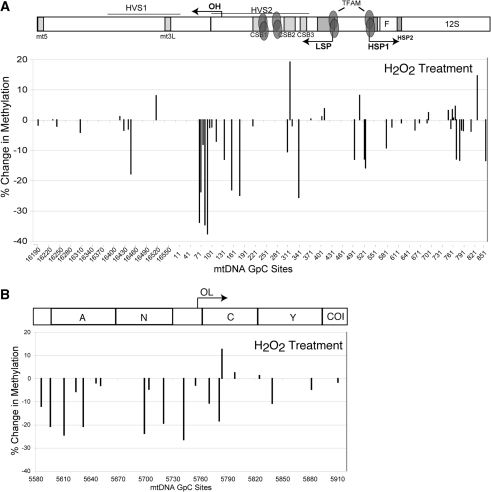
mtDNA nucleoids are transiently more compact during oxidative stress.
In order to investigate the dynamics of mtDNA nucleoids remodeling in response to environmental factors, we exposed mito-M.CviPI-HA cells to oxidative stress (H2O2). To test the DNA methyltransferase accessibility to mtDNA in the presence of ROS, Mito-M.CviPI-HA expressing cells were treated with 200 µM H2O2 for 1 h and subsequently induced for mito-M.CviPI-HA expression for 1 h. We observed an overall decrease in mtDNA methylation after 1 h induction, suggesting a higher compact state of the mtDNA in the presence of H2O2 (Figure 10). This result suggests that the mtDNA nucleoid becomes more compact under oxidative stress.
Figure 10.
mtDNA methylation levels are decreased when cells are exposed to H2O2. (A) Quantification of relative changes in methylation in cells treated with H2O2 in the following selected regions: NCR (A) and OL (B).
DISCUSSION
By targeting DNA methyltransferases to the mitochondria, we have developed a novel tool to identify protected regions in the mtDNA in living cells. In contrast, increased methylation frequency observed at some sites might indicate increased accessibility associated to DNA bending and looping formation caused by remodeling factors such as TFAM. When compared to the average mtDNA methylation level after induction of mito-M.CvipI, the most protected GpC sites identified are located upstream the mt3L region, in the LSP promoter (mostly in HeLa), in the HSP and nearby regions, mTERF1 binding site (TERM) and in the OL region. Our analyses indicate that mtDNA is not methylated in the cells utilized under standard culture conditions. Shmookler Reis et al reported that ∼2–5% mtDNA are methylated at CCGG regions (28), but there was no indication of GpC methylation, and these findings from the early 80’s, have not been substantiated. Nonetheless, extreme low percentage of methylation (<5%) would not be detected by the approach employed.
The protection of the TERM region was much more robust than other mtDNA regions, indicating that this site is persistently occupied. Transcripts initiated at the HSP promoter (PH1) site give rise to the mitochondrial rRNAs and are terminated by mTERF1 binding to TERM. Such transcripts are initiated ∼50–100 times more frequently than overlapping transcripts initiated from HSP promoter PH2 that gives rise to the full-length heavy-strand transcript (29). mTERF1 has been shown to bind TERM with high affinity and to be necessary for mitochondrial rDNA looping required for maintaining the high levels of rRNA transcription (30). Our results show that TERM occupancy is very high even under conditions of TFAM knockdown/overexpression or recovery from mtDNA depletion, conditions that would be expected to lead to changes in mtDNA activity and packaging. It is unclear how transcription proceeds through this region to generate most of the H-strand-coded RNAs. It has been reported that mTERF1 binds OL with moderate affinity (20) and it may be responsible for the methylation protection observed in the region.
Methylation patterns were conserved in different types of cells and in models based on different expression systems. The mTERF1 binding site showed a similar degree of protection in all cell lines analyzed; however, the TFAM binding site to the LSP showed the highest degree of protection in HeLa cells, possibly because of more frequent transcription initiation events.
Methylation levels were influenced by factors known to initiate changes in the nucleoid environment. Cells recovering from mtDNA depletion induced by EtBr treatment, had increased overall methylation levels indicating a mtDNA less packed with proteins during active mtDNA replication. Increased mtDNA accessibility can also be explained by the decreased TFAM: mtDNA ratio during mtDNA repletion. It has been shown that TFAM levels, which also get depleted when mtDNA is not present, recovered to normal levels more slowly than mtDNA after transient depletion by EtBr treatment (31). Since TFAM is the most abundant nucleoid protein involved in mtDNA packaging, a decrease in the TFAM: mtDNA ratio could result in a less compact mtDNA. This result supports the idea that the ratio of TFAM: mtDNA might be a major regulator of mtDNA activity (i.e. expression/replication), and may vary according to cell’s metabolic demands. TFAM has been shown to be developmentally regulated in Xenopus oocytes (32). During oogenesis, TFAM levels increase considerably as the oocyte matures, a stage during which mtDNA activity is reduced. In yeast the ratio of the TFAM homologue Abf2 to mtDNA is reduced under respiratory growth conditions; Conversely, Abf2 levels increase under glucose repression and amino-acid starvation, making the mtDNA more compact (24).
In this study, the effects of TFAM levels on mtDNA accessibility were analyzed and we observed a decrease in mtDNA methylation when TFAM was either down- or up-regulated. When TFAM levels were reduced by siRNA, mtDNA levels were also decreased. However, it is possible that in this situation other nucleoid components may still be present at their own steady-state levels. This may make the overall structure of nucleoids less accessible to the methyltransferase. When TFAM is overexpressed, the mtDNA binding sites in the promoter region may become saturated with TFAM, suppressing methyltransferase accessibility. This result supports the concept that the steady state levels of TFAM is not enough to fully saturate the mtDNA, otherwise overexpression of TFAM would not affect methyltransferase accessibility. It has been demonstrated that excess TFAM can also lead to mtDNA depletion by causing a shift towards a slow replication mode (10). We believe that excess TFAM may result in overcompaction of the nucleoid and consequently making it difficult for other replication factors to access the promoter region. Although moderate increases of TFAM levels can stimulate transcription and replication, high concentrations of TFAM have the opposite inhibitory effect (9). The decrease in mtDNA methylation was less prominent at other regions, suggesting that TFAM is not equally distributed across the mtDNA, not even when it is present at high concentrations. At high TFAM levels, the preferential binding specificity to the promoter region may be maintained.
ATAD3 depletion was also accompanied by a reduction in mtDNA methylation which might be associated with changes in mtDNA topology to a more relaxed state. Increased open circular forms of mtDNA accompanied by loss of PicoGreen staining was also reported when prohibitin 1(PHB1), another peripheral nucleoid protein was knocked down (33). Although the exact function of ATAD3 remains unknown, it was proposed to be a peripheral nucleoid protein, which along with PHB1 and HSP60, surrounds the nucleoid core (25). This suggestion was based on the fact that ATAD3 has been found in nucleoid preparations (5) and it has a high binding affinity to D-loop structures in vitro (21). However, others have demonstrated that ATAD3 fails to crosslink to mtDNA and it cannot bind mtDNA in vivo (25). Whether ATAD3 interacts with mtDNA directly or not, it may still have a role in structural remodeling of mtDNA. More work will be required to test how peripheral nucleoid proteins play a role in mtDNA organization and stability.
Finally, we used the in vivo methylation approach to look for changes in nucleoid structure triggered by oxidative stress. mtDNA is quite sensitive to reactive oxygen species (ROS), since it does not have a complex chromatin organization. TFAM may play an important role in protecting the mtDNA against oxidative damage and as well as repairing the mtDNA. It has been demonstrated that sub-lethal doses of H2O2 cause a drastic increase in the ratio of TFAM protein per mtDNA (34). In addition, Yoshida et al. (35) demonstrated that TFAM preferentially binds oxidatively damaged DNA in vitro. The increased binding affinity of TFAM to oxidative damaged DNA, may modulate the mtDNA structure to a more compact state than normal. Accordingly, we observed an overall decreased mtDNA methylation when cells were exposed to H2O2 for 1 h. A potential caveat, is that because M.CvipI recognizes GpC sites, it is possible that methylation could be inhibited by the presence of 8-hydroxy-2′-deoxyguanosine (8-oxodG). However, it has been demonstrated that 8-oxoG content in circular mtDNA is relatively low after oxidative stress (36,37). Because 8-oxodG can be removed from the mtDNA by a base excision repair process involving 8-oxoguanine DNA glycosylase (OGG1) (38), the recruitment of repair enzymes may reduce the accessibility of mtDNA to M.CviPI. It is also possible that nucleoids remodel into a more compact state during oxidative stress to provide a more insulated environment for mtDNA. As mentioned above, the changes we observed in mtDNA methylation during oxidative stress may be regulated by interaction between TFAM and mtDNA. Other nucleoid factors may also play a role in protecting mtDNA against ROS. Recently it has been shown that SOD2 (manganese superoxide dismutase) is an integral nucleoid protein that could confer mtDNA protection against ROS (39). In addition, the mismatch repair enzyme, MYH is recruited to oxidized DNA in order to excise A/8-oxodG mismatch (24). All these factors could contribute to a more packed mtDNA that is less accessible to the methyltransferase. More detailed analysis of mtDNA interactions with protein factors will help elucidate the functional dynamics of mitochondrial nucleoid structures.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
PHS grants EY010804, NS041777 and CA085700. Pre-doctoral fellowship from the American Heart Association (to A.R.). Funding for open access charge: PHS grant NS041777.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors are grateful to Dr Michael P. Kladde (University of Florida) for the M.CviPI construct.
REFERENCES
- 1.Garrido N, Griparic L, Jokitalo E, Wartiovaara J, van der Bliek AM, Spelbrink JN. Composition and dynamics of human mitochondrial nucleoids. Mol. Biol. Cell. 2003;14:1583–1596. doi: 10.1091/mbc.E02-07-0399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alam TI, Kanki T, Muta T, Ukaji K, Abe Y, Nakayama H, Takio K, Hamasaki N, Kang D. Human mitochondrial DNA is packaged with TFAM. Nucleic Acids Res. 2003;31:1640–1645. doi: 10.1093/nar/gkg251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Legros F, Malka F, Frachon P, Lombes A, Rojo M. Organization and dynamics of human mitochondrial DNA. J. Cell Sci. 2004;117:2653–2662. doi: 10.1242/jcs.01134. [DOI] [PubMed] [Google Scholar]
- 4.Nunnari J, Marshall WF, Straight A, Murray A, Sedat JW, Walter P. Mitochondrial transmission during mating in Saccharomyces cerevisiae is determined by mitochondrial fusion and fission and the intramitochondrial segregation of mitochondrial DNA. Mol. Biol. Cell. 1997;8:1233–1242. doi: 10.1091/mbc.8.7.1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang Y, Bogenhagen DF. Human mitochondrial DNA nucleoids are linked to protein folding machinery and metabolic enzymes at the mitochondrial inner membrane. J. Biol. Chem. 2006;281:25791–25802. doi: 10.1074/jbc.M604501200. [DOI] [PubMed] [Google Scholar]
- 6.Kanki T, Nakayama H, Sasaki N, Takio K, Alam TI, Hamasaki N, Kang D. Mitochondrial nucleoid and transcription factor A. Ann. NY Acad. Sci. 2004;1011:61–68. doi: 10.1007/978-3-662-41088-2_7. [DOI] [PubMed] [Google Scholar]
- 7.Fisher RP, Lisowsky T, Parisi MA, Clayton DA. DNA wrapping and bending by a mitochondrial high mobility group-like transcriptional activator protein. J. Biol. Chem. 1992;267:3358–3367. [PubMed] [Google Scholar]
- 8.Larsson NG, Wang J, Wilhelmsson H, Oldfors A, Rustin P, Lewandoski M, Barsh GS, Clayton DA. Mitochondrial transcription factor A is necessary for mtDNA maintenance and embryogenesis in mice. Nat. Genet. 1998;18:231–236. doi: 10.1038/ng0398-231. [DOI] [PubMed] [Google Scholar]
- 9.Maniura-Weber K, Goffart S, Garstka HL, Montoya J, Wiesner RJ. Transient overexpression of mitochondrial transcription factor A (TFAM) is sufficient to stimulate mitochondrial DNA transcription, but not sufficient to increase mtDNA copy number in cultured cells. Nucleic Acids Res. 2004;32:6015–6027. doi: 10.1093/nar/gkh921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pohjoismaki JL, Wanrooij S, Hyvarinen AK, Goffart S, Holt IJ, Spelbrink JN, Jacobs HT. Alterations to the expression level of mitochondrial transcription factor A, TFAM, modify the mode of mitochondrial DNA replication in cultured human cells. Nucleic Acids Res. 2006;34:5815–5828. doi: 10.1093/nar/gkl703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fisher RP, Clayton DA. Purification and characterization of human mitochondrial transcription factor 1. Mol. Cell Biol. 1988;8:3496–3509. doi: 10.1128/mcb.8.8.3496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Antoshechkin I, Bogenhagen DF, Mastrangelo IA. The HMG-box mitochondrial transcription factor xl-mtTFA binds DNA as a tetramer to activate bidirectional transcription. Embo J. 1997;16:3198–3206. doi: 10.1093/emboj/16.11.3198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Takamatsu C, Umeda S, Ohsato T, Ohno T, Abe Y, Fukuoh A, Shinagawa H, Hamasaki N, Kang D. Regulation of mitochondrial D-loops by transcription factor A and single-stranded DNA-binding protein. EMBO Rep. 2002;3:451–456. doi: 10.1093/embo-reports/kvf099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kaufman BA, Durisic N, Mativetsky JM, Costantino S, Hancock MA, Grutter P, Shoubridge EA. The mitochondrial transcription factor TFAM coordinates the assembly of multiple DNA molecules into nucleoid-like structures. Mol. Biol. Cell. 2007;18:3225–3236. doi: 10.1091/mbc.E07-05-0404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wiesner RJ, Zsurka G, Kunz WS. Mitochondrial DNA damage and the aging process: facts and imaginations. Free Radical Res. 2006;40:1284–1294. doi: 10.1080/10715760600913168. [DOI] [PubMed] [Google Scholar]
- 16.Cotney J, Wang Z, Shadel GS. Relative abundance of the human mitochondrial transcription system and distinct roles for h-mtTFB1 and h-mtTFB2 in mitochondrial biogenesis and gene expression. Nucleic Acids Res. 2007;35:4042–4054. doi: 10.1093/nar/gkm424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jessen WJ, Dhasarathy A, Hoose SA, Carvin CD, Risinger AL, Kladde MP. Mapping chromatin structure in vivo using DNA methyltransferases. Methods. 2004;33:68–80. doi: 10.1016/j.ymeth.2003.10.025. [DOI] [PubMed] [Google Scholar]
- 18.van Steensel B, Delrow J, Henikoff S. Chromatin profiling using targeted DNA adenine methyltransferase. Nat. Genet. 2001;27:304–308. doi: 10.1038/85871. [DOI] [PubMed] [Google Scholar]
- 19.Carvin CD, Dhasarathy A, Friesenhahn LB, Jessen WJ, Kladde MP. Targeted cytosine methylation for in vivo detection of protein-DNA interactions. Proc. Natl Acad. Sci. USA. 2003;100:7743–7748. doi: 10.1073/pnas.1332672100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hyvarinen AK, Pohjoismaki JL, Reyes A, Wanrooij S, Yasukawa T, Karhunen PJ, Spelbrink JN, Holt IJ, Jacobs HT. The mitochondrial transcription termination factor mTERF modulates replication pausing in human mitochondrial DNA. Nucleic Acids Res. 2007;35:6458–6474. doi: 10.1093/nar/gkm676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.He J, Mao CC, Reyes A, Sembongi H, Di Re M, Granycome C, Clippingdale AB, Fearnley IM, Harbour M, Robinson AJ, et al. The AAA + protein ATAD3 has displacement loop binding properties and is involved in mitochondrial nucleoid organization. J. Cell Biol. 2007;176:141–146. doi: 10.1083/jcb.200609158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Frommer M, McDonald LE, Millar DS, Collis CM, Watt F, Grigg GW, Molloy PL, Paul CL. A genomic sequencing protocol that yields a positive display of 5-methylcytosine residues in individual DNA strands. Proc. Natl Acad. Sci. USA. 1992;89:1827–1831. doi: 10.1073/pnas.89.5.1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Leakey TI, Zielinski J, Siegfried RN, Siegel ER, Fan CY, Cooney CA. A simple algorithm for quantifying DNA methylation levels on multiple independent CpG sites in bisulfite genomic sequencing electropherograms. Nucleic Acids Res. 2008;36:e64. doi: 10.1093/nar/gkn210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xiong Z, Laird PW. COBRA: a sensitive and quantitative DNA methylation assay. Nucleic Acids Res. 1997;25:2532–2534. doi: 10.1093/nar/25.12.2532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Suzuki H, Hosokawa Y, Nishikimi M, Ozawa T. Existence of common homologous elements in the transcriptional regulatory regions of human nuclear genes and mitochondrial gene for the oxidative phosphorylation system. J. Biol. Chem. 1991;266:2333–2338. [PubMed] [Google Scholar]
- 26.Kucej M, Kucejova B, Subramanian R, Chen XJ, Butow RA. Mitochondrial nucleoids undergo remodeling in response to metabolic cues. J. Cell Sci. 2008;121:1861–1868. doi: 10.1242/jcs.028605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bogenhagen DF, Rousseau D, Burke S. The layered structure of human mitochondrial DNA nucleoids. J. Biol. Chem. 2008;283:3665–3675. doi: 10.1074/jbc.M708444200. [DOI] [PubMed] [Google Scholar]
- 28.Shmookler Reis RJ, Goldstein S. Mitochondrial DNA in mortal and immortal human cells. Genome number, integrity, and methylation. J. Biol. Chem. 1983;258:9078–9085. [PubMed] [Google Scholar]
- 29.Gelfand R, Attardi G. Synthesis and turnover of mitochondrial ribonucleic acid in HeLa cells: the mature ribosomal and messenger ribonucleic acid species are metabolically unstable. Mol. Cell Biol. 1981;1:497–511. doi: 10.1128/mcb.1.6.497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Martin M, Cho J, Cesare AJ, Griffith JD, Attardi G. Termination factor-mediated DNA loop between termination and initiation sites drives mitochondrial rRNA synthesis. Cell. 2005;123:1227–1240. doi: 10.1016/j.cell.2005.09.040. [DOI] [PubMed] [Google Scholar]
- 31.Seidel-Rogol BL, Shadel GS. Modulation of mitochondrial transcription in response to mtDNA depletion and repletion in HeLa cells. Nucleic Acids Res. 2002;30:1929–1934. doi: 10.1093/nar/30.9.1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shen EL, Bogenhagen DF. Developmentally-regulated packaging of mitochondrial DNA by the HMG-box protein mtTFA during Xenopus oogenesis. Nucleic Acids Res. 2001;29:2822–2828. doi: 10.1093/nar/29.13.2822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kasashima K, Sumitani M, Satoh M, Endo H. Human prohibitin 1 maintains the organization and stability of the mitochondrial nucleoids. Exp. Cell Res. 2008;314:988–996. doi: 10.1016/j.yexcr.2008.01.005. [DOI] [PubMed] [Google Scholar]
- 34.Noack H, Bednarek T, Heidler J, Ladig R, Holtz J, Szibor M. TFAM-dependent and independent dynamics of mtDNA levels in C2C12 myoblasts caused by redox stress. Biochim. Biophys. Acta. 2006;1760:141–150. doi: 10.1016/j.bbagen.2005.12.007. [DOI] [PubMed] [Google Scholar]
- 35.Yoshida Y, Izumi H, Ise T, Uramoto H, Torigoe T, Ishiguchi H, Murakami T, Tanabe M, Nakayama Y, Itoh H, et al. Human mitochondrial transcription factor A binds preferentially to oxidatively damaged DNA. Biochem. Biophys. Res. Commun. 2002;295:945–951. doi: 10.1016/s0006-291x(02)00757-x. [DOI] [PubMed] [Google Scholar]
- 36.Shokolenko I, Venediktova N, Bochkareva A, Wilson GL, Alexeyev MF. Oxidative stress induces degradation of mitochondrial DNA. Nucleic Acids Res. 2009;37:2539–2548. doi: 10.1093/nar/gkp100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Suter M, Richter C. Fragmented mitochondrial DNA is the predominant carrier of oxidized DNA bases. Biochemistry. 1999;38:459–464. doi: 10.1021/bi9811922. [DOI] [PubMed] [Google Scholar]
- 38.de Souza-Pinto NC, Eide L, Hogue BA, Thybo T, Stevnsner T, Seeberg E, Klungland A, Bohr VA. Repair of 8-oxodeoxyguanosine lesions in mitochondrial DNA depends on the oxoguanine DNA glycosylase (OGG1) gene and 8-oxoguanine accumulates in the mitochondrial dna of OGG1-defective mice. Cancer Res. 2001;61:5378–5381. [PubMed] [Google Scholar]
- 39.Kienhofer J, Haussler DJ, Ruckelshausen F, Muessig E, Weber K, Pimentel D, Ullrich V, Burkle A, Bachschmid MM. Association of mitochondrial antioxidant enzymes with mitochondrial DNA as integral nucleoid constituents. FASEB J. 2009;23:2034–2044. doi: 10.1096/fj.08-113571. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.